Figure 7
- ID
- ZDB-FIG-211216-48
- Publication
- Gomes et al., 2021 - New Findings on LMO7 Transcripts, Proteins and Regulatory Regions in Human and Vertebrate Model Organisms and the Intracellular Distribution in Skeletal Muscle Cells
- Other Figures
- All Figure Page
- Back to All Figure Page
|
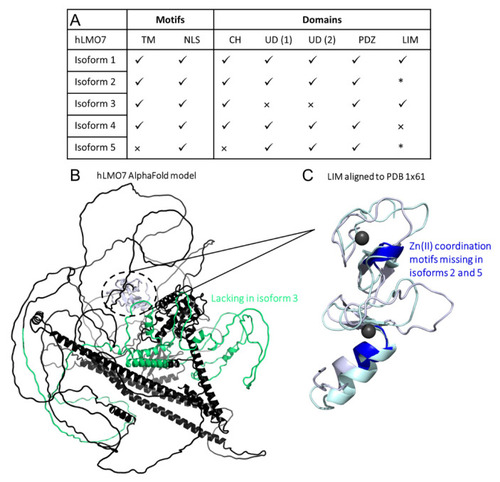
Comparison of domains organization of the five isoforms from human LMO7. (A) Summary of results from the primary structure alignment among the hLMO7 isoforms. √, presence. ×, absence. * lacks Zn(II) binding motifs. TM, transmembrane α-helix. NLS, nuclear localization sequence. CH, calponin homology domain. UD, unknown domain DUF4757. (B) hLMO7 Alpha Fold-predicted structural model showing the region missing in isoform 3 (green) and the LIM domain (white) which is only conserved in isoforms 1 and 3. (C) Superimposition of the LIM domain of hLMO7 (obtained by AlphaFold; white ribbon representation) and the LIM domain of TRIP6 (PDB 1X61; pale green ribbon representation), which is the closest structural analog reported by I-TASSER [42]. Two zinc ions (black spheres) are coordinated in the fingers. Regions highlighted in dark blue are missing in isoforms 2 and 5. Furthermore, isoform 4 does not have the full LIM domain. |