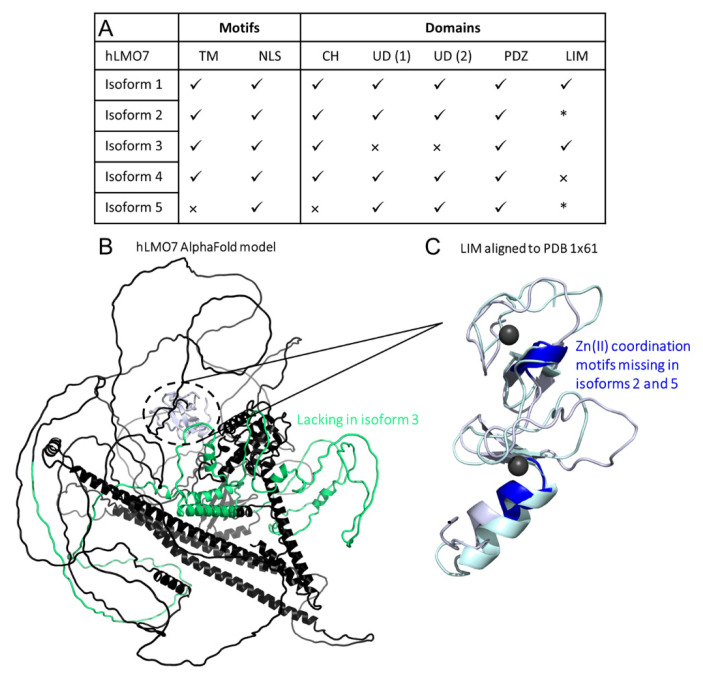
Figure 7 Comparison of domains organization of the five isoforms from human LMO7. (A) Summary of results from the primary structure alignment among the hLMO7 isoforms. √, presence. ×, absence. * lacks Zn(II) binding motifs. TM, transmembrane α-helix. NLS, nuclear localization sequence. CH, calponin homology domain. UD, unknown domain DUF4757. (B) hLMO7 Alpha Fold-predicted structural model showing the region missing in isoform 3 (green) and the LIM domain (white) which is only conserved in isoforms 1 and 3. (C) Superimposition of the LIM domain of hLMO7 (obtained by AlphaFold; white ribbon representation) and the LIM domain of TRIP6 (PDB 1X61; pale green ribbon representation), which is the closest structural analog reported by I-TASSER [42]. Two zinc ions (black spheres) are coordinated in the fingers. Regions highlighted in dark blue are missing in isoforms 2 and 5. Furthermore, isoform 4 does not have the full LIM domain.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Int. J. Mol. Sci.