Figure 3
- ID
- ZDB-FIG-211216-44
- Publication
- Gomes et al., 2021 - New Findings on LMO7 Transcripts, Proteins and Regulatory Regions in Human and Vertebrate Model Organisms and the Intracellular Distribution in Skeletal Muscle Cells
- Other Figures
- All Figure Page
- Back to All Figure Page
|
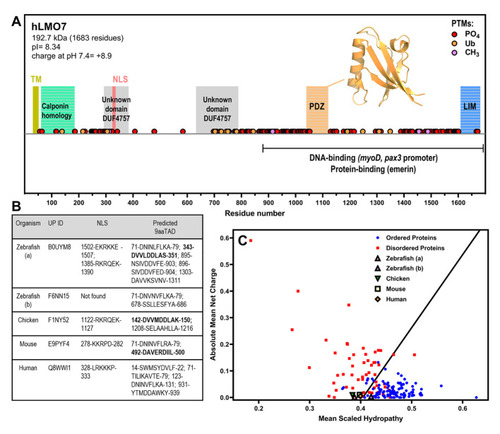
LMO7 is a multidomain protein that has a charge–hydropathy characteristic of proteins enriched in intrinsic disorder and contains nuclear localization sequence(s) and putative nine-amino-acid transactivation motifs. (A) Schematic organization of human LMO7 with the domains, motifs and post-translational modifications (PTMs) indicated along sequence. The C-terminal domain mediates interaction with DNA promoters and protein partners. TM, transmembrane α-helix. NLS, nuclear localization sequence. PO4, phosphorylation. Ub, ubiquitination. CH3, methylation. The inset shows the X-ray crystallographic structure of the PDZ domain (PDB 2eaq). (B) LMO7 from different species were analyzed by NLSdb (third column, https://rostlab.org/services/nlsdb/, 20 November 2021) and 9aaTAD (fourth column, https://www.med.muni.cz/9aaTAD/). Perfect matches of 9aaTAD are highlighted in bold whereas other annotated motifs correspond to 92% matches. Corresponding UniProt identifiers (UP ID) are shown in the second column. (C) Predictor of Natural Disordered Regions (PONDR; http://www.pondr.com/) analysis of charge–hydropathy (CH plot). Well-folded proteins indicated as blue circles and disordered proteins as red squares. LMO7 from the different species is located in the plot region of disordered proteins apart from isoform b of zebrafish. |