Figure 1—figure supplement 1.
- ID
- ZDB-FIG-201209-30
- Publication
- Segebarth et al., 2020 - On the objectivity, reliability, and validity of deep learning enabled bioimage analyses
- Other Figures
-
- Figure 1—figure supplement 1.
- Figure 1—figure supplement 1.
- Figure 1—figure supplement 2.
- Figure 2—figure supplement 1.
- Figure 2—figure supplement 1.
- Figure 2—figure supplement 2.
- Figure 2—figure supplement 3.
- Figure 2—figure supplement 4.
- Figure 3—figure supplement 1—source data 1.
- Figure 3—figure supplement 1—source data 1.
- Figure 4—source data 2.
- Figure 5—figure supplement 1.
- Figure 5—figure supplement 1.
- Figure 5—figure supplement 2.
- Figure 5—figure supplement 3.
- Figure 5—figure supplement 4.
- Figure 5—figure supplement 5.
- All Figure Page
- Back to All Figure Page
|
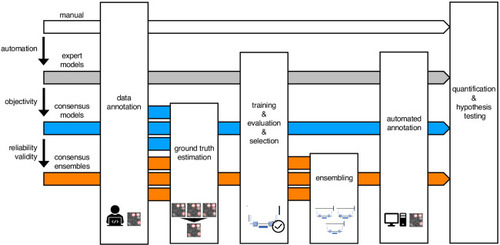
Schematic illustration of bioimage analysis strategies and corresponding hypotheses. Four bioimage analysis strategies are depicted. Manual (white) refers to manual, heuristic fluorescent feature annotation by a human expert. The three DL-based strategies for automatized fluorescent feature annotation are based on expert models (gray), consensus models (blue) and consensus ensembles (orange). For all DL-based strategies, a representative subset of microscopy images is annotated by human experts. Here, we depict labels of cFOS-positive nuclei and the corresponding annotations (pink). These annotations are used in either individual training datasets (gray: expert models) or pooled in a single training dataset by means of ground truth estimation from the expert annotations (blue: consensus models, orange: consensus ensembles). Next, deep learning models are trained on the training dataset and evaluated on a holdout validation dataset. Subsequently, the predictions of individual models (gray and blue) or model ensembles (orange) are used to compute binary segmentation masks for the entire bioimage dataset. Based on these fluorescent feature segmentations, quantification and statistical analyses are performed. The expert model strategy enables the automation of a manual analysis. To mitigate the bias from subjective feature annotations in the expert model strategy, we introduce the consensus model strategy. Finally, the consensus ensembles alleviate the random effects in the training procedure and seek to ensure reliability and eventually, validity. |