Fig. 1
- ID
- ZDB-FIG-100223-39
- Publication
- Raeker et al., 2010 - Targeted deletion of the zebrafish obscurin A RhoGEF domain affects heart, skeletal muscle and brain development
- Other Figures
- All Figure Page
- Back to All Figure Page
|
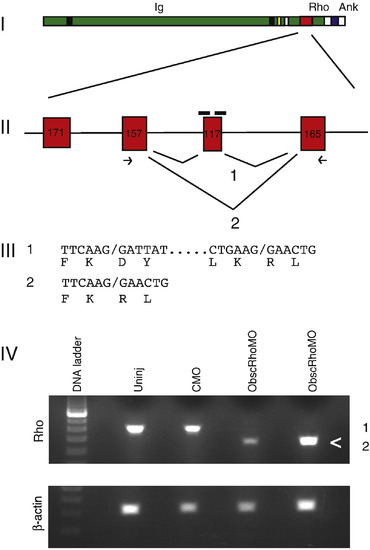
Targeting of the obscurin A RhoGEF domain. (I) Diagram of obscurin A demonstrating the location of the tandem immunoglobulin-like (Ig) domains, the RhoGEF domain (Rho) and the ankyrin binding (Ank) domains. (II) Diagram of the exons encoding the double homology (DH) domain of the RhoGEF module. A 117-bp exon encoding for part of the GTPase activation site of the RhoGEF domain was targeted for induced exon skipping by the injection of antisense morpholinos (black bars) directed at the splice donor and acceptor sites for that exon. (III) Splice product 1 represents the normal splicing. The morpholinos are frequently able to block normal splicing of the targeted exon resulting in the splice product 2. PCR primers (arrows) from the adjacent exons were used to detect the splice products. Both products were cloned and sequenced and the nucleotide and predicted amino acid sequences displayed. (IV) RNA from 48 hpf embryos was isolated from obscurin A RhoGEF (ObscRhoMO) morphant, uninjected (Uninj) control morpholino-injected (CMO) embryos. Note that the band (<) corresponding to splice product 2 is only present in the RNA isolated from the morphant embryos. |
Reprinted from Developmental Biology, 337(2), Raeker, M.O., Bieniek, A.N., Ryan, A.S., Tsai, H.J., Zahn, K.M., and Russell, M.W., Targeted deletion of the zebrafish obscurin A RhoGEF domain affects heart, skeletal muscle and brain development, 432-443, Copyright (2010) with permission from Elsevier. Full text @ Dev. Biol.