- Title
-
A brain-specific angiogenic mechanism enabled by tip cell specialization
- Authors
- Schevenels, G., Cabochette, P., America, M., Vandenborne, A., De Grande, L., Guenther, S., He, L., Dieu, M., Christou, B., Vermeersch, M., Germano, R.F.V., Perez-Morga, D., Renard, P., Martin, M., Vanlandewijck, M., Betsholtz, C., Vanhollebeke, B.
- Source
- Full text @ Nature
|
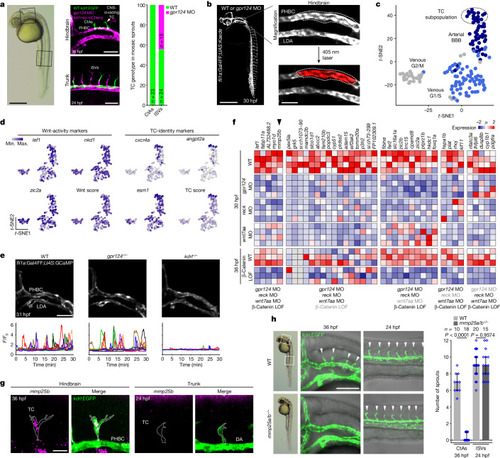
Mmp25 as a regulator of brain-specific angiogenesis. |
|
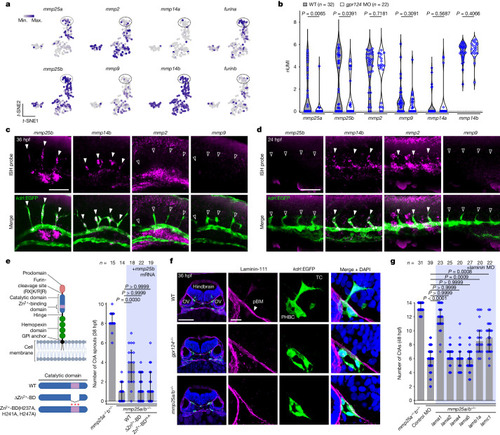
TCs require a specialized MMP repertoire to migrate across the pial basement membrane. |
|
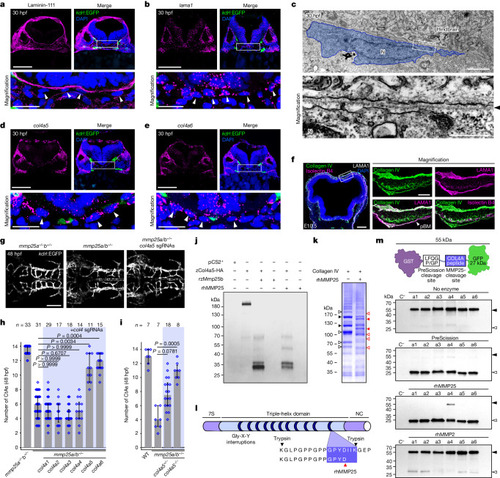
Mmp25 cleaves meningeal fibroblast-derived Col4a5/6. |
|
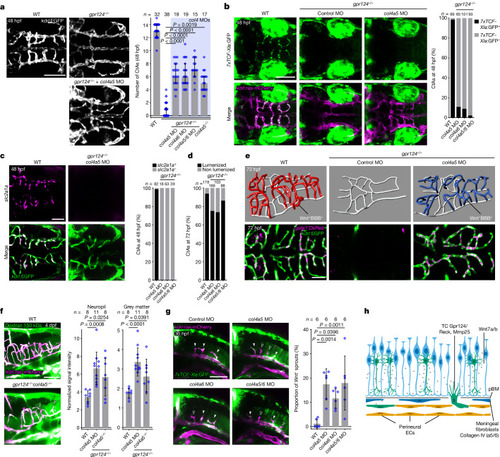
Col4a5/6 inactivation unlocks the quality control of brain angiogenesis by Wnt–β-catenin signalling. |