- Title
-
Long wavelength-sensing cones of zebrafish retina exhibit multiple layers of transcriptional heterogeneity
- Authors
- Farre, A.A., Sun, C., Starostik, M.R., Hunter, S.S., English, M.A., Duncan, A., Santhanam, A., Shihabeddin, E., O'Brien, J., Swaroop, A., Stenkamp, D.L.
- Source
- Full text @ Front. Cell. Neurosci.
|
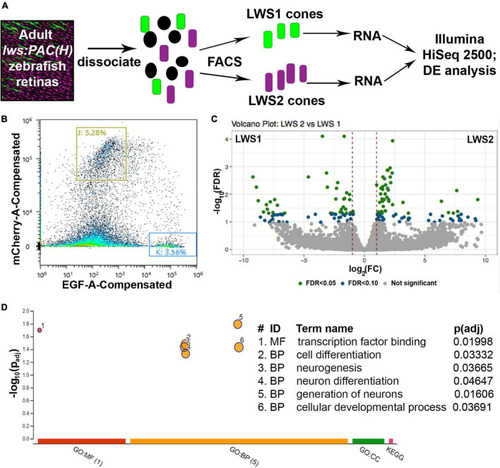
Comparative transcriptome analysis of LWS1 vs. LWS2 cones using FACS followed by bulk RNA-Seq. |
|
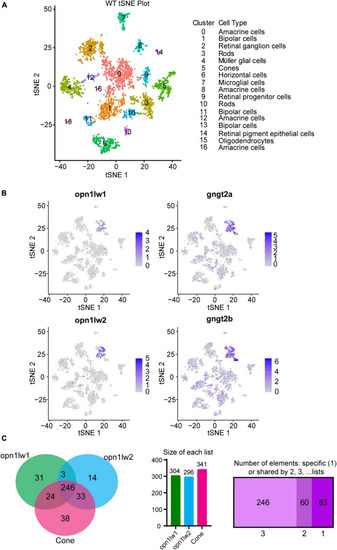
Single cell RNA-Seq and interrogation for transcripts enriched in LWS1 vs. LWS2 cones. |
|
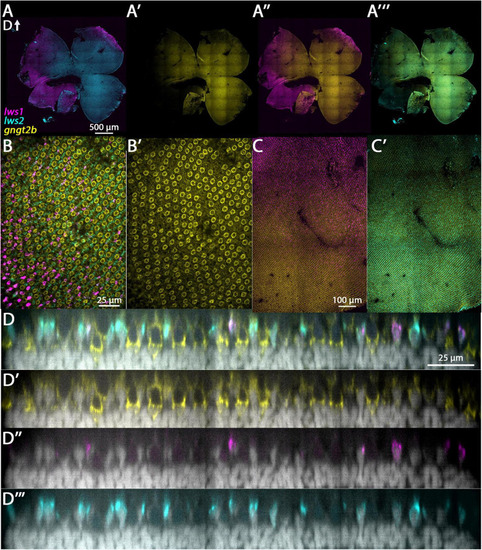
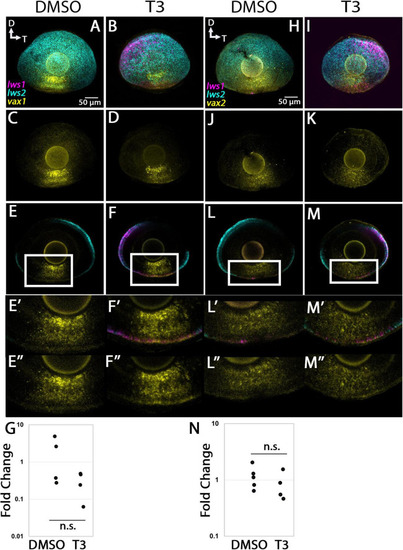
Expression of |
|
Expression of |
|
Expression of |
|
Expression of |
|
Expression of |
|
Expression of |
|
Expression of |
|
Expression of |
|
Expression of |
|
Expression of |