- Title
-
dnmt1 function is required to maintain retinal stem cells within the ciliary marginal zone of the zebrafish eye
- Authors
- Angileri, K.M., Gross, J.M.
- Source
- Full text @ Sci. Rep.
|
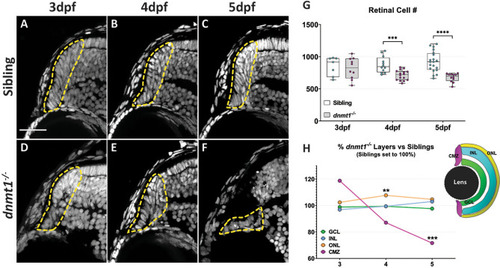
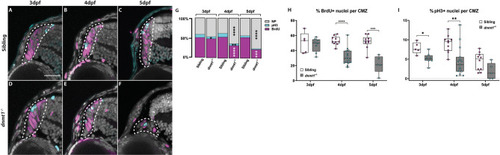
Disruption of dnmt1 function results in CMZ defects. |
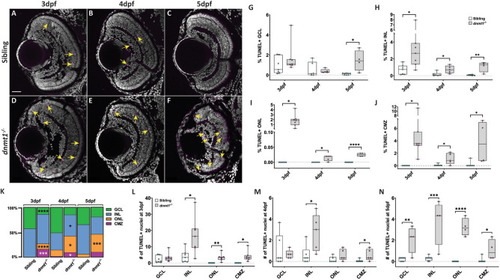
|
Cell death is elevated in the |
|
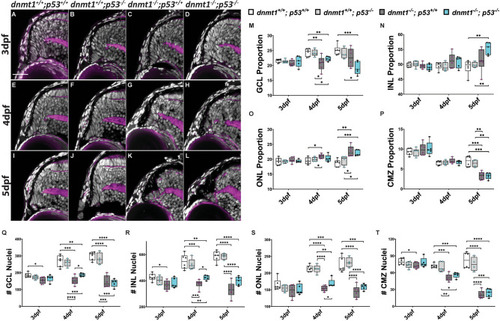
Loss of p53 function does not rescue the |
|
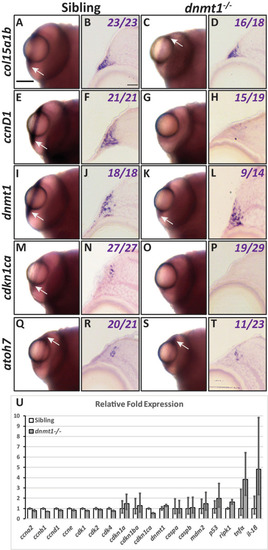
dnmt1 is required to maintain RSC gene expression. Gene expression shown in whole mount ( |
|
RSCs require dnmt1 function to maintain proliferation. |
|
Neurons produced by |
|
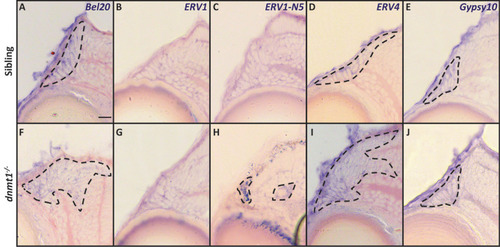
Loss of dnmt1 function results in misregulation of retroelement expression. |
|
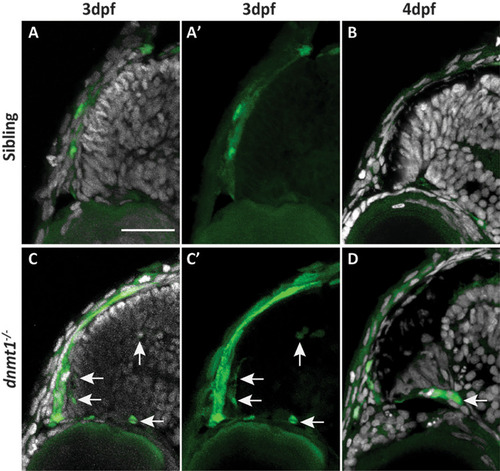
RSCs require dnmt1 function to repress L1RE3-EGFP transposition. |
|
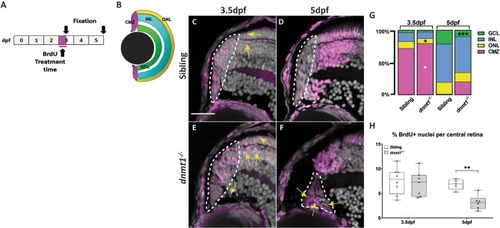
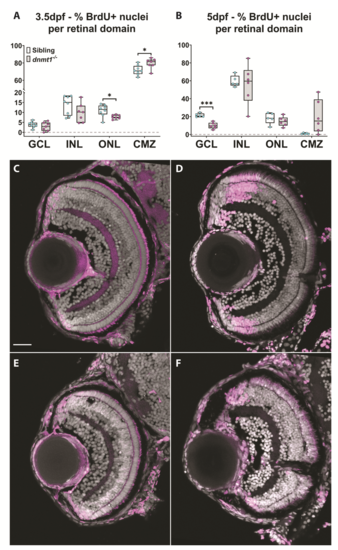
dnmt1-deficient RSCs fail to incorporate into the neural retina. A. Data points collected of the proportion of cells labeled with BrdU in each retinal domain at 3.5dpf divided by the number of total BrdU+ cells. B. Data points collected of the proportion of cells labeled with BrdU in each layer at 5dpf divided by the number of total BrdU+ cells. Sibling controls = white bars; dnmt1-/- = gray bars. *p <0.05, ***p <0.0005. C-F. Transverse sections of sibling (C-D) and dnmt1-/- (E-F) retinae at 3.5dpf (C,E) and 5dpf (D,F). Nuclei labeled with DAPI (gray). BrdU+ cells (magenta). Dorsal is up in all images. Scale bar (C) = 30 μm. All images taken at the same magnification. |
|
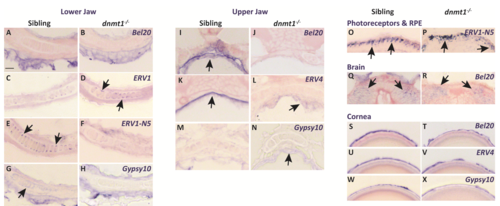
Loss of dnmt1 function results in misregulation of LTR RE expression across numerous tissues. A-X. Transverse cryosections of larvae analyzed by in situ hybridization. All sibling and dnmt1-/- larvae are 4dpf. A-H. Expression of indicated REs within the lower jaw. I-N. Expression of indicated REs within the upper jaw. O-P. Expression of ERV1-N5 LTR in photoreceptors (O; sibling) and the RPE (P; dnmt1-/-). Q-R. Expression of Bel20 LTR within the brain. S-X. Expression of indicated REs within the cornea. Arrows delineate expression changes of specified REs. Scale bar (A) = 10 μm. All images taken at the same magnification. RPE = retinal pigmented epithelium. |
|
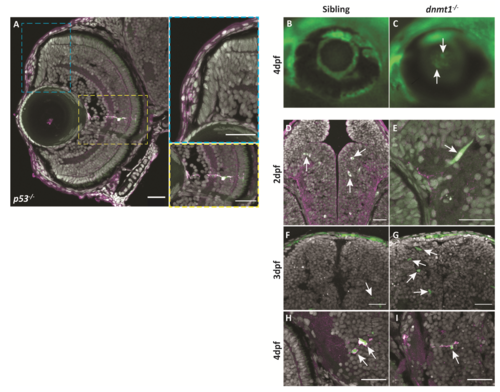
L1RE3-EGFP transgene expression is more prominent in dnmt1-/- larvae. A-A’’. Transverse section of Tg(CMV:Hsa.L1RE3,EGFP,myl7:EGFP;p53-/-) larvae at 4dpf. A. L1RE3-EGFP+ retinal cells labeled with endogenous EGFP. Cyan (A‘) and yellow (A’‘) dotted boxes indicate magnified images to the right of panel A. A’. Magnified image of Tg(CMV:Hsa.L1RE3,EGFP,myl7:EGFP;p53-/-) CMZ showing no expression of the L1RE3-EGFP transgene. A’’. Magnified image of retinal neuron expressing L1RE3-EGFP transgene. B-I. All images are taken from Tg(CMV:Hsa.L1RE3,EGFP,myl7:EGFP) larvae that are either dnmt1+/+ (B,D,F,H) or dnmt1-/- (C,E,G,I). B-C. Whole-mount images of 4dpf eyes demonstrating L1RE3-EGFP transgene activation seen through the lens of dnmt1-/- larvae and not siblings. D,F,H. Sibling larvae expressing the L1RE3-EGFP transgene in the brain. E,G,I. dnmt1-/- larvae expressing the L1RE3-EGFP transgene in the brain. Nuclei labeled with DAPI (gray). Endogenous L1RE3-activated EGFP labeled in green. EGFP antibody is magenta. Scale bars: 30 μm in all images. Dorsal is up in all images. |