- Title
-
Itch Is Required for Lateral Line Development in Zebrafish
- Authors
- Angers, A., Drapeau, P.
- Source
- Full text @ PLoS One
|
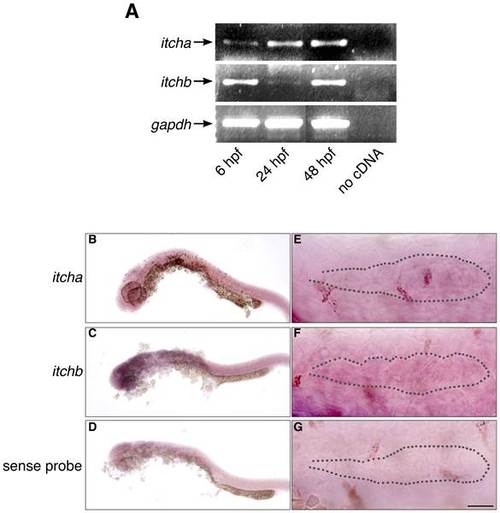
Expression of itch genes in zebrafish embryos. (A) RT-PCR experiment showing early expression of both itcha and itchb genes. RNA was extracted from embryos at approximately 6, 24 and 48 hpf. cDNA was obtained from each stage using 1 µg total RNA. GAPDH was used as an internal control for cDNA amplification. (B–D) Weak, general staining of embryos at 26 hpf revealed with DIG-labelled probes complementary to itcha (B) or itchb (C) or sense sequence (D) as a control. (E–G) The primordium of 26 hpf embryos revealed with DIG-labelled probes complementary to itcha (E) or itchb (F) or sense sequence (G) as a control. Position of the primordium is underlined with a dotted line. Expression of itch genes is general, and itchb is expressed throughout the primordium. Scale bar: (B–D) 250 µm, (E–G) 25 µm. |
|
itch knockdown impairs early zebrafish development. (A) Splice junction MOs against itcha and itchb efficiently reduced their respective but not opposite mRNAs. RNA was extracted from 48 hpf control embryos or embryos injected with MOs against itcha (MoA) or itchb (MoB). After reverse-transcription, cDNAs from each group were amplified with PCR primers specific for itcha, itchb or GAPDH as a control for 35 cycles. Marked reduction of itcha and itchb was obtained after injection of the appropriate but not the other MO. (B–I) Morphology of embryos injected with MOs targeting itchb (MoB) showed general growth failure, already visible at 24 hpf (B,C) and persisting through to 96 hpf (H,I) as compared to control embryos (CTRL). Scale bar: 500 µm. (J) Total length of control embryos or embryos injected with MOs against itchb (MoB) was measured from the tip of the head to the tip of the tail muscle. Two-factor analysis of variance indicates no significant interaction of injection with age. Pairwise comparison was performed between control and MoB injected embryos at each time point. The number of embryos measured is indicated at the bottom of the graph. ** represents statistical significance at p < 0.01. (K-N) Morphology of control embryos (CTRL), itcha knockdown embryos (MoA), itchb knockdown (MoB) or itcha and itchb knockdown embryos (MoA+B) at 48 hpf. itcha knockdown embryos were slightly delayed, but were morphologically intact as compared to itchb or itcha plus itchb knockdown embryos. (O) Global growth of injected embryos was assessed by total length measurements at 72 hpf. itcha and itchb knockdown embryos were significantly smaller than control embryos, and itchb knockdown were smaller than itcha as assessed by Kruskal-Wallis one-way ANOVA combined with Dunn′s method of group comparison with a significant threshold fixed at p < 0.01 (**). itcha plus itchb knockdown embryos were not significantly smaller than itchb knockdown embryos (n.s.). (P) Growth defect of itchb knockdown was partially rescued with injection of in vitro transcribed human ITCH mRNA. Kruskal-Wallis one-way ANOVA combined with Dunn′s method of comparison established no-significant differences between control embryos (CTRL) and embryos injected with ITCH mRNA alone (mRNA), whereas the total length was significantly reduced in both itchb MO-injected embryos (MoB) and embryos injected with a combination of itchb MO and ITCH mRNA (Rescue). Total length was significantly larger in the Rescue group than in the MoB group, indicating partial rescue. The number of embryos measured is indicated at the bottom of the graphs. Each graph summarizes at least four different experiments.(**) indicates p < 0.01. |
|
itchb is involved in pLL primordium migration and lateral line development. (A–E) Posterior lateral line at 48 hpf cldnb:gfp in embryos injected with vehicle (CTRL), in vitro-transcribed human ITCH mRNA (mRNA), MOs against itcha (MoA), MOs against itchb (MoB) or MOs against itchb and in vitro-transcribed human ITCH mRNA (Rescue). There was a marked reduction in the number of neuromasts in the pLL after itchb knockdown (D), but not in itcha knockdown, although the primordium had not yet reached the end of the tail at this time point (C). The itchb knockdown effect was partially rescued by injection of human ITCH mRNA (E). (F–I) Time-lapse confocal microscopy on 26–30 hpf cldnb:gfp embryos showed that the pLL primordium migration was slowed in itchb knockdown embryos (MoB) compared to vehicle (CTRL), human ITCH mRNA-injected (mRNA) or rescued embryos. (N) The number of neuromasts in the posterior lateral-line was counted after staining with DiAsp in 72 hpf WT embryos injected with vehicle (CTRL), in vitro-transcribed human ITCH mRNA (mRNA), MOs against itcha (MoA), MOs against itchb (MoB) or MOs against itchb and in vitro-transcribed human ITCH mRNA (Rescue). Whereas the number of neuromasts in the pLL was markedly reduced after injection of MOs against itchb, injection of in vitro transcribed ITCH mRNA alone or of MOs against itcha had no effect. The MOs phenotype was rescued by co-injection of ITCH mRNA in itchb knockdown. The number of embryos for each group is indicated at the bottom of the graph. ** represents statistical significance at p < 0.01. n.s. = non-significant. (J–M) Morphology of the migrating primordium in 26 hpf embryos (CTRL), itcha knockdown embryos (MoA) and itchb knockdown embryos (MoB). Cells at the tip of the primordium of MoB embryos were slightly disorganized, but the typical rosette formation occurred normally. (O) Migration of the pLL primordium was quantified by measuring the distance between the primordium leading edge in the first frame (t=0) compared to the last frame (t=60) as shown in F–I (doted line). The number of embryos counted or imaged is indicated at the bottom of the graph. Each graph summarizes four different experiments. ** represents statistical significance at p <0.01. n.s. = non-significant. EGFP fluorescence is presented as reversed black and white to facilitate visualization in all micrographs. Scale bars: (A–E) 34 µm, (F–I) 22 µm, (L–M) 40 µm. |
|
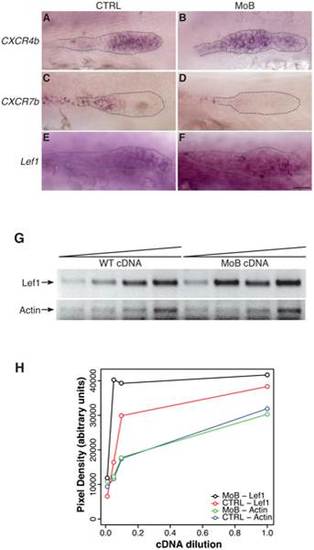
Primordium patterning is altered in itchb knocked-down embryos. (A–F) RNA in situ hybridization of factors required for primordium patterning in control embryos (vehicle-injected siblings) (CTRL, left panels) and MOs-injected embryos (MoB, right panels) at 30 hpf. cldnb:gfp embryos were used to ensure that the primordium had migrated pass the 10th somite before fixation. (A,B) cxcr4b expression was limited to the leading half of the primordium in control embryos, but extended throughout the primordium after itchb knockdown. (C,D) cxcr7b was limited to the trailing end of control embryos, and almost completely excluded from the primordium in itchb knockdown. (E,F) lef1 was expressed in the leading edge of control embryos. Darker staining indicated higher expression in embryos injected with MOs against itchb (MoB), but leading edge expression was maintained. Scale bar: 25 µm. (G) Semi-quantitative RT-PCR showing enhanced expression of lef1 in embryos injected with MOs against itchb (MoB) as compared to control embryos (WT). Amplification of the actin gene was used as an internal control. The image was inverted to facilitate quantification. (H) Densitometry measurements from the gel presented in G. This is representative of three different experiments. EXPRESSION / LABELING:
|