- Title
-
Scube2 mediates Hedgehog signalling in the zebrafish embryo
- Authors
- Hollway, G.E., Maule, J., Gautier, P., Evans, T.M., Keenan, D.G., Lohs, C., Fischer, D., Wicking, C., and Currie, P.D.
- Source
- Full text @ Dev. Biol.
|
Homozygous you mutants lack HH sensitive cell fates. Panels A–D youty97 mutants show a reduction in ptc expression within the somites and neural tube. Wild type (WT) embryos at the 10-somite (A) or 22-somite stages (C) show robust expression of ptc compared to similar stage homozygous youty97 mutant embryos panels B and D. A–B dorsal views, panels C, D lateral views, anterior to the left. Panels E–L comparative phenotype of different you alleles and intra allelic compounds at 26 hpf. Lateral views anterior to the left. (E) Wildtype myotomes demonstrate a distinct chevron shape and the presence of a horizontal myosepta (arrows). (F) youty97 homozygotes exhibit u-shaped somites and lack a horizontal myosepta. (G) ptc is strongly expressed in wildtype myotomes at this stage (26 hpf). (H) ptc expression is reduced in homozygous youty97 mutants. (I) Homozygous youtm146c mutants exhibit a much less severe alteration in myotomal shape. (J) Intra allelic homozygote compounds between youty97 and youtm146c exhibit a myotomal phenotype intermediate between the homozygote phenotype of the two separate alleles. (K) Myotomal ptc expression is indistinguishable from wild type in homozygous youtm146c mutant embryos. (L) ptc expression in youtm146c/ty97 allelic compound homozygote embryos is reduced compared to youtm146chomozygotes. (M, N) youty97/syudel double mutant embryos exhibit an additive phenotype. (M) 26 somite syudel homozygous mutant embryos exhibit a more severe reduction in somitic ptc staining than homozygous youty97 mutants. (N) Ptc staining is reduced to barely detectable levels within similarly staged double mutant youty97 and syudel embryos. (O, P) myoD expression at the 15-somite stage, specifically within the adaxial cells, is down regulated compared to wildtype (O) in youty97 homozygote embryos (P), arrows indicate gaps in adaxial expression. Panels Q–T engrailed mRNA expression in the pioneer cells at the 26-somite stage is severely down regulated as compared to wildtype (Q, S) within homozygous youty97 mutants (R, T). Arrowheads in panels Q and R mark the midbrain–hindbrain expression of eng which is unaffected within you homozygote mutant embryos. (U, V) Floor plate expression of axial (hnf3β) is reduced or absent compared to wildtype (U) within youty97 homozygote mutant embryos (V). (W, X) At 26 somites, slow myosin heavy chain gene expression is severely reduced compared to wild type embryos (W) within youty97 homozygous mutants embryos (X). EXPRESSION / LABELING:
|
|
Slow muscle formation is disrupted in homozygous youmutants. (A–H) Slow myosin heavy chain protein (SMHC) is reduced compared to wild type panels A, C, E, G in youty97 homozygote mutant embryos panels B, D, F, H at 10-somite (A, B) and 26-somite stages (C–H). Panels E, F anterior somites, (G, H) posterior, tail extension somites. (E–H) 3D confocal projections of left sided myotomes. (I, J) At 26 somites, Prox1 protein, a specific marker of slow muscle cells, is reduced or absent compared to wild type (I) within the myotome of youty97 homozygous embryos (J). A–B dorsal confocal sections, at the level of the notochord, anterior to the left. (C–J) Lateral views anterior to the left. EXPRESSION / LABELING:
|
|
Identification of Scube2 as the gene mutated at the you locus. (A) Positional cloning of the you locus. A critical region on linkage group 7 (LG7) was defined by simple sequence repeat marker (SSR) mapping and marker z11119 was found to be non-recombinant in 1400 meioses. A probe derived from the non-repetitive portion of z11119 detected PAC2D2. Further screening using PAC end cloning isolated BAC9A3. These two clones were shotgun sequenced and this sequence subsequently used to identify contiguous stretches of sequence in the emerging zebrafish genome sequence (ctg 08658, ctg 01991, ctg 09688,). The scube2 open reading frame was identified within this sequence, and the full genomic structure of the gene identified with reference to identified cDNA sequences. Intron sequences between exons 15 and 17 could not be defined due to gaps in the assembled genomic sequence. Arrow at exon 14 indicates the site of the identified stop codon in the youty97 allele. Arrow at exon 5 indicates the site of the identified stop codon in the youtm146c allele with the blue lines indicating the alternative splicing detected for this exon. (B) A stop codon is produced by mutation of a C to a T at the first base of codon Q644 and segregates with the youty97 phenotype. A stop codon in produced by mutation of a G to a T at the first base of codon E164 and segregates with the youtm146c phenotype. (C) Bootstrap phylogenetic analysis of all known Scube proteins, reveals that the you gene encodes a zebrafish protein orthologous to mammalian Scube2. The tree is rooted with human matrillin 2, the most closely related protein outside the Scube family. Abbreviations: hs, Homo sapiens; mm, Mus musculus; gg, Gallus gallus; dr, Danio rerio; tn, Tetraodon nigroviridis. (D) Structural homology of Cubilin and Scube2. The region sufficient for Cubilin binding to Megalin is bracketed. The position of the stop codon produced by the youty97 and youtm146c mutations is indicated by red arrows. (E) The EGF repeats of Scube2 and Cubilin share specific amino acid residues that define a separate Calcium binding EGF repeat class. Pan alignment of 73 seed sequences representative of all 5329 EGF domains present within PFAM and all EGF domains of the calcium binding class. This analysis reveals that specific EGF repeats (blue box) of zebrafish (dr) Scube2 (2, 3, 5, 6, 7, 9) share closest identity and specific residues (red arrow heads) with specific EGF repeats of human proteins, Cubilin (cbn), the LDL receptor (ldlr) and the LDLR related protein LRP-5. (F) Table of EGF domain homologies. A matrix of highest level homologies for the nine zebrafish Scube2 EGF domains reveals a separate sub-class of EGF containing proteins. Human Cubulin (cbn) and other endocytic receptors of the LDL receptor class (LRP5 and LDLR) share a high and distinct level of homology. The EGF repeats of human Attractin (attrn), which are not within this class, are included for comparison. (G) Western blot showing that zebrafish Scube2 is secreted from Scube2 transfected cells. Media from myc-tagged Scube2 transfected cells (Scube2) contains two secreted forms of zebrafish Scube2 protein, the larger at a similar molecular weight predicted by the full ORF and from similar experiments utilising the mammalian Scube proteins. These proteins are absent from media harvested from untransfected cells (CON). Marker sizes in kDa are shown. EXPRESSION / LABELING:
|
|
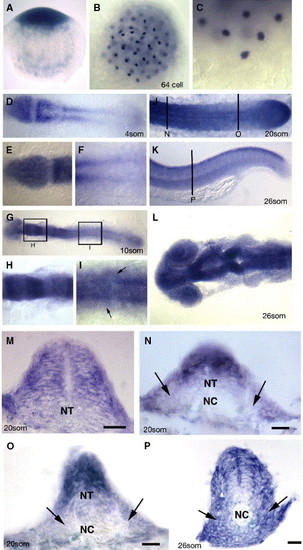
Embryonic expression of zebrafish scube2. (A–P) Expression of the gene (blue) encoded by the you locus as revealed by in situ hybridisation. A. you mRNA is deposited maternally as evidenced by the detection of mRNA in the one cell stage embryo. Lateral view, dorsal to the top. (B) Maternally deposited you mRNA is localised to discrete regions in cleavage stage embryos, as evidenced by in situ hybridisation on this 64 cell stage embryo. Dorsal view. (C) High magnification view of the most ventral cells of the same embryo as in (B). Lateral view dorsal to the top. (D) 4-somite stage embryos show strong expression of you mRNA in the anterior neural plate, newly formed somites and PSM. (E) High magnification of you expression in the neural plate at the same stage as in (D). (F) High magnification at the same stage as in (D) of expression within PSM. (G) Expression of you mRNA at the 10-somite stage reveals a modulation of expression within different regions of the hindbrain (high magnification view in panel H), and continued expression within the neural tube and PSM. Expression within the posterior neural tube is also evident at this stage making expression within the neural epithelium global (High magnification view in (I), at the dorsal ventral-level of the notochord, arrows indicate expression within somitic mesoderm cells). (J) Expression at the 20-somite stage continues in the neural tube, somites and PSM, including robust expression within the tail bud (arrow). (D–J dorsal views anterior to the left). (K) At the end of somitogenesis (26-somite stage), expression within the posterior neural tube and myotome is down regulated with the exception of the dorsal most aspect of the neural tube. Lateral view anterior to the left. Vertical line denotes level of the section (P). (L) At 26 somites, the anterior neural tube expression remains high, and modulates within specific neuronal regions, dorsal view anterior to the left. (M) Transverse section of the anterior neural tube of 20-somite stage embryo revealing expression throughout the entire neural epithelium. Panels N–P transverse section from the locations and stages indicated in panels J and K, Arrows demark the somitic expression of zebrafish scube2. Scale bars 20 μM. PSM—presomitic mesoderm, NT—neural tube, NC—notochord. |
|
Non-cell autonomous somitic requirement for the gene encoded by the you locus in myotomal cell fate determination. (A–M) Transplantation of wildtype blastula cells into homozygous mutant youty97 host embryos. (A) Rhodamine dextran labelled donor cells (red) transplanted into a homozygous youty97 host embryo targeted to posterior, tail extension somites, rescues formation of both muscle pioneer cells (arrows) and slow MyHC expression (green). (B) Slow MyHC protein (SMyHC) expression (green) is rescued by transplantation of wild type cells. (C) Position of transplanted clones is revealed by Rhodamine dextran. (D–G) Cross sections at the level of yolk extension myotomes revealing that transplantation of wild type cells (red) in the notochord of a homozygous youty97 mutant does not rescue muscle pioneer and slow muscle formation (green). (D) Slow MyHC in green, wildtype donor cells in red. (E) Wildtype donor cells (red) only. (F) Slow MyHC staining (green) only. (G) A wild type embryo sectioned at a similar level stained for slow MyHC (green) is shown for comparison. The presence of muscle pioneers is indicated by arrows. (H–J) Transplantation of wildtype donor cells (green) into the most dorsal aspect of the neural tube of a homozygous mutant youty97 mutant embryos cannot rescue the formation of engrailed (eng) expressing muscle cells, as revealed by in situ hybridisation with an antisense probe against the eng1 gene, a marker of muscle pioneer formation, utilising the Fast Red (Roche) fluorescent substrate (red). (H) Eng-expressing cells (red) and wildtype donor cells (green). (I) Eng staining (red) only. (J) wildtype donor cells (green) only. (K–M) High magnification view of the region boxed in panel H. The arrows in panel K mark the site of the donor cells. (L) Note the lack of eng expression in somites adjacent to the transplanted cells (M). (N–Y) Transplantation of homozygous youty97 mutant blastula cells into wild type host embryos. (N–P) Dorsal views anterior to the left. A single mutant cell transplanted into the region of the yolk extension somites of a wildtype host can differentiate as an eng (red) expressing muscle pioneer cell, revealing a non-autonomous action of the you gene product. Panel N. Overlay of Eng staining (red) and donor staining (green). (O) Eng staining (red) only. (P) Donor cell staining (green) only. (Q–Y) Transplantation of mutant cells (green) into the neural tube of wild type host embryos does not disrupt muscle pioneer formation (red). (Q–V) Low magnification lateral views, anterior to the left, of yolk extension somites in transplanted hosts showing a high level of chimerism. Vertical lines denote the corresponding sections, panels W–Y. (Q and T) Overlay of eng staining (red) and donor staining (green). Panels R and U. eng staining (red) only. Panels S and V. donor cell staining (green) only. Panel W. Cross-section revealing the contribution of mutant donor cells to engrailed positive muscle pioneer cells (arrows). Note also that transplanted cells in the dorsal neural tube fail to interfere with muscle pioneer formation. (X, Y) Engrailed positive myocytes (arrows) still form even when mutant cells comprise the entire dorsal aspect of the neural tube. Scale bars, 20 μM. WT—wildtype, MUT—youty97 homozygous mutant. |
|
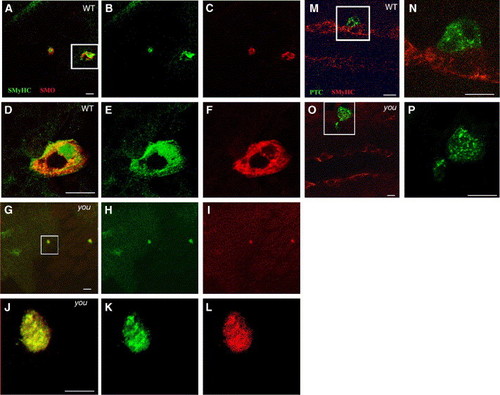
Scube2 is not required for the cytoplasmic localisation of Patched or Smoothened. (A–F) Constitutively active Smoh mutant M1 induces Slow MyHC expression cell autonomously within the PSM of wild type and youty97 mutant embryos and is localised predominately intracellularly within a 15-somite stage embryo. Oblique, dorsal confocal sections at the level of the notochord anterior to the top. (A) Slow MyHC (SMyHC) expression (green) within the adaxial cells and ectopically within activated Smoh M1 (red) clones. (B) SMyHC alone. (C) Smoh protein expression alone. (D–F) High magnification views of the single cell expressing activated Smoh boxed in panel A. (D) Slow MyHC (SMyHC) expression (green) within the activated Smoh M1 (red) clones, revealing a predominately intracellular localisation within induced clones. (E) SMyHc alone. (F) Smoh protein expression alone. (G–L) Induction of slow MyHC expression and Smoh localisation are unaffected when mosaic Smoh M1 clones are induced within a 15-somite stage homozygous youty97 mutant embryo. (G) Mosaic Smoh M1 expression (red) induces slow MyHC expression (green) in a youty97 homozygote embryo. (H) SMyHc alone. (I) Smoh protein expression alone. (J–L) High magnification views of the single cell, boxed in panel G, expressing activated Smoh and activated Smoh M1 (red). (J) Slow MyHC (SMyHC) expression (green) within the activated Smoh M1 (red) clones. (K) SMyHc alone. (L) Smoh protein expression alone, revealing a predominately intracellular localisation of Smoh protein within expressing clones, similar to clones induced in wildtype embryos. (M, N) Ptc distribution within a wildtype 15-somite stage embryo, monitored using C-terminally fused GFP is constitutively intracellular, regardless of localisation of Ptc expressing clones within the PSM. Dorsal confocal sections at the level of the notochord, anterior to the left. (M) SMyHC (red) denotes the position of the adaxial cells with a single Ptc (Green) expressing cell localised immediately lateral to the adaxial cells. (N) High magnification view of the region boxed in panel M revealing Ptc is localised primarily to intracellular vesicular structures suggestive of endosomal elements, consistent with previous observations (Incardona et al., 2002 and Evans et al., 2003). (O, P) Localisation of PTC within intracellular foci does is not altered by expressing PTC-GFP within a youty97 homozygous mutant embryo. (P) High magnification view of the region boxed in panel O. Panels O and P are similar stages and views to those shown in panels M, N. Scale bars, 20 μm. |
|
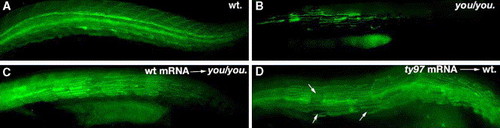
Scube2 mRNA rescues the defects present in youty97 mutants. (A) Uninjected wildtype sibling embryo stained for Slow MyHC (green). (B) Uninjected youty97 sibling embryo. (C) youty97 embryo injected with full-length scube2 mRNA showing a moderate rescue phenotype. (D) Injection of the mRNA encoding the truncated form of Scube2 (youty97 mutant form) into wildtype results in proportion of embryos that exhibit a mild loss of slow muscle cells that is often restricted to individual myotomes (arrows). |
Reprinted from Developmental Biology, 294(1), Hollway, G.E., Maule, J., Gautier, P., Evans, T.M., Keenan, D.G., Lohs, C., Fischer, D., Wicking, C., and Currie, P.D., Scube2 mediates Hedgehog signalling in the zebrafish embryo, 104-118, Copyright (2006) with permission from Elsevier. Full text @ Dev. Biol.