- Title
-
Nuclear Pores Regulate Muscle Development and Maintenance by Assembling a Localized Mef2C Complex
- Authors
- Raices, M., Bukata, L., Sakuma, S., Borlido, J., Hernandez, L.S., Hart, D.O., D'Angelo, M.A.
- Source
- Full text @ Dev. Cell
|
Nup210 Is Required for Skeletal Muscle Development (A) Zebrafish one-cell embryos injected with Control or Nup210 morpholinos (MO) were imaged at different times of development. Asterisks show cardiac edema. (B) The length, height (dorsal-ventral length), and angle of somite #20 were quantified in control and Nup210-depleted animals stained with the F59 antibody. (C) Zebrafish embryos injected with control or Nup210 MOs were stained with F59 and phalloidin at 48 hpf. Images represent the maximal projection of multiple z stacks. (D) Embryos were co-injected with control or Nup210 morpholinos and GST mRNA, or a Nup210 mRNA resistant to the morpholino (Nup210r) and muscle was analyzed at 48 hpf. (E) Rescue experiments from (D) were quantified by measuring somite angle. (F) Muscle structure in Nup210 CRISPR knockouts was analyzed at 48 hpf by staining with F59 and phalloidin. Bar plots represent mean ± SEM, n ≥ 3 replicates. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p<0.001, ∗∗∗∗p ≤ 0.0001, two-tailed Student's t test. Morpholino depletions were performed with n ≥ 50 embryos. Ten to 20 embryos were examined by immunofluorescence and quantified in n ≥ 3 independent experiments. Scale bars, 50 μm. See also Figure S1. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Nup210 Depletion Does Not Affect Early Muscle Development (A) Whole-mount in situ hybridization against the muscle markers Pax3, Myf5, and MyoD were performed at different times of development in zebrafish embryos injected with control or Nup210 morpholinos. (B) Slow muscle was stained with F59 at different times of development in control and Nup210-depleted animals and analyzed by confocal microscopy. (C) Control or Nup210 morphants were incubated with cyclopamine during development to inhibit the Sonic Hedgehog-dependent formation of early embryonic slow muscle. Addition of new muscle fibers during muscle growth was analyzed by F59 staining at 48 hpf. Morpholino depletions were performed with n ≥ 50 embryos. Ten to 20 embryos were examined by immunofluorescence and quantified in n ≥ 3 independent experiments. Scale bars, 50 μm. See also Figure S1. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Nup210 Depletion Inhibits Myofibrillogenesis and Results in Muscle Cell Death (A) Thin filament structure in control and Nup210 morphants was analyzed by phalloidin staining at different times of development. (B) Tail muscle was stained for apoptotic death with an anti-activated caspase-3 antibody in control and Nup210-depleted animals. (C) Cross-sections of zebrafish animals at 96 hpf were stained with the slow muscle marker F59 (green), phalloidin (red), and DAPI (blue) (n = 6–10 embryos). Representative images of n ≥ 3 independent experiments. n = 10–20 embryos unless otherwise specified. Scale bars, 50 μm. |
|
Nup210 Interacts with and Regulates Muscle Development through Trip6 (A) The expression levels of several sarcomeric and muscle contraction genes in Nup210-depleted C2C12 myotubes was extracted from D'Angelo et al. (2012). (B) Nup210 was immunoprecipitated (IP) from wild-type or FLAG-Trip6-expressing myotubes and the presence of Trip6 was determined using anti-Trip6 or anti-FLAG antibodies. Nup210 is expressed at low levels and is not detected in the input. Lamin A and Hsp90 were analyzed in Nup210 immunoprecipitates to determine the specificity of Trip6 binding (right panel). IgG, immunoglobulin G. (C) The association of zebrafish Nup210 C-terminal domain fused to GST with different in vitro translated HA-tagged LIM-domain proteins was analyzed using pull-down assays. (D) Embryos were co-injected with control or Nup210 morpholinos and mRNA for GST, full-length Nup210, or Nup210ΔCT, and muscle structure was analyzed 48 hpf with the F59 antibody. Myotome angle was used to quantify muscle alterations (n = 10–20 embryos, performed in triplicate). (E) C2C12 cells were infected with lentiviruses carrying control, Nup210, or Trip6 shRNAs, selected and induced to differentiate. Differentiated myotubes were stained for myosin heavy chain (MHC) at 72 hr post differentiation. Nuclei were stained with Hoechst. (F) Zebrafish embryos were injected with control or Trip6 morpholinos and slow muscle was stained with F59 at 48 hpf (left panel). Muscle alterations were quantified by measuring myotome angle (right panel). n = 10–20 embryos, performed in triplicate. (G) Control, Nup210, or Trip6 morpholinos (top panel) and subphenotypic concentrations of Nup210, Trip6, or both morpholinos (bottom panel) were injected into one-cell zebrafish embryos and slow muscle was stained at 48 hpf. Bar plots represent mean ± SEM, n ≥ 3 replicates. ∗p ≤ 0.05. Scale bar, 50 μm. See also Figure S2. |
|
Nup210 Interacts with and Recruits Mef2C to NPCs (A) Nup210 and Mef2C immunofluorescence in differentiated C2C12 cells (day 4). MHC was used as a marker for differentiated myotubes. Nuclei were stained with Hoechst. Arrowheads show quiescent myoblasts with non-detectable Nup210 or Mef2C. (B) C2C12 cells were infected with lentiviruses carrying control and Mef2C shRNAs, induced to differentiate and stained at 72 hr after differentiation with MHC. (C) Nup210 was immunoprecipitated from wild-type or FLAG-Mef2C expressing C2C12 myotubes, and the presence of Mef2C was determined with a pan-MEF2 antibody (top panel) or an anti-FLAG (bottom panel) antibody. Mef2C-specific band was confirmed by shRNA depletion (Figure S2B). Asterisk shows the potential Mef2A and Mef2D bands recognized by the pan-MEF2 antibody as described by the manufacturer. The specificity of these bands was not confirmed in this study. Nup210 is expressed at low levels in myotubes and not detected in the input. (D) Differentiated C2C12 cells were analyzed by immunofluorescence against Nup210 and Mef2C (IF), and proximity ligation assays (PLA). PLA shows the interaction of Mef2C with Nup210 at the nuclear periphery (red dots). Representative images (n ≥ 3). Scale bars, 5 μm. (E) Zebrafish embryos were co-injected with Nup210 morpholinos plus GST or Mef2Cα mRNAs and slow muscle was stained at 48 hpf. (F) Rescue experiments from (E) were quantified by measuring somite angle (n = 10–20 embryos, performed in triplicate). (G) Zebrafish embryos were injected with control or Nup210 morpholinos alone or in combination with Mef2Cα RNA, Pom121 MO, or Pom121 MO + Mef2Cα RNA. Slow muscle was analyzed 48 hpf. (H) Quantification of myotome angle from (G) (n = 10–20 embryos, performed in triplicate). (I) Control or Nup210 morpholino-depleted embryos were stained at 48 hpf for Mef2C (red dots) and F-actin (magenta). Nuclei were stained with Hoechst (blue). (J) The localization of Mef2C in control or Nup210-depleted myotubes was analyzed by immunofluorescence. Top panel shows the downregulation of Nup210 in shRNA-depleted myotubes. Scale bar, 20 μm. Bar plots represent mean ± SEM, n ≥ 3 replicates. ∗p ≤ 0.05. Scale bars represent 50 μm unless otherwise specified. See also Figure S2. |
|
Nup210 role in skeletal muscle growth. Related to Figures 1 and 2. (A) U2OS cells were transfected with a vector expressing V5-tagged Zebrafish Nup210. Cells were stained using the V5 antibody and the NPC antibody mAb414. Top panel shows a cross-section of the nucleus. Bottom panel shows NPCs on the nuclear surface. Zebrafish Nup210 localizes to NPCs. n ≥ 3 (B) Zebrafish one-cell embryos were injected with control or two different Nup210 morpholinos and slow muscle fibers were stained 48 hours post-fertilization with the F59 antibody. Representative image of a maximum projection of 30-40 sections. (C) Schematic illustration of the myogenic waves in Zebrafish based on Barresi et al (Barresi et al., 2001). During segmentation slow muscle fibers form in the myotomes from adaxial-derived precursors (red, first myogenic wave). When segmentation is complete, at ~24 hpf, new muscle fibers form a different set of muscle precursors and are added to the dorsal and ventral side of the myotome in a process known as stratified hyperplasia (green, second myogenic wave). Sonic Hedgehog (Shh) is required for the first, but not the second, myogenic wave. Thus, the Shh inhibitor cyclopamine inhibits the formation of the early development muscle fibers (red) but not the myofibers that are added post-segmentation (green). (D) Illustration showing the experimental approach to study the role of Nup210 in postsegmentation muscle growth using cyclopamine. (E) Control or Nup210 morphants were stained 48 hpf with the slow muscle F59 antibody and the number of myofibers in myotome 20 were quantified. Bar plots represent mean ± SEM, ∗∗∗∗p ≤ 0.0001, two-tailed Student's t test, n ≥ 3 replicates. Morpholino depletions were performed with n ≥ 50 embryos. n = 10-20 embryos from at least three independent experiments were examined by immufluorescence and quantified. |
|
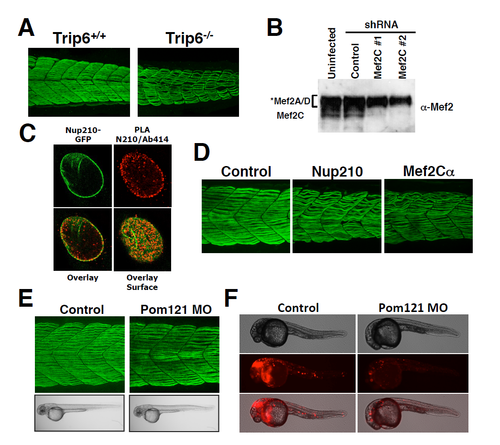
Trip6, Mef2C and Pom121 depletion in Zebrafish embryos. Related to Figures 4 and 5. (A) Wild-type or Trip6 CRISPR-generated mutants were stained with the F59 antibody at 48 hpf. Slow muscle was imaged by confocal microscopy. Representative image of a maximum projection of 30-40 sections. n ≥ 10 embryos. (B) C2C12 cells were infected at 36 hours post-differentiation with lentivirus carrying control or MEF2C-specific shRNAs. Cells were collected 72 hours post-infection and Mef2C depletion was analyzed by western blot using an anti-Mef2 antibody. According to the manufacturer anti-Mef2 recognizes Mef2A, C and D. Western blots show the specific Mef2C protein band. The specificity for Mef2A and D was not confirmed (C) The interaction of the nucleoproin Nup210 (green) with nucleoporins recognized by the mAb414 antibody, were analyzed by proximity ligation assays. PLA interactions are shown in red. (C) MEF2Cα was depleted during Zebrafish development using specific morpholinos previously characterized (Hinits and Hughes, 2007). Slow muscle was stained at 48 hpf and compared to Control or Nup210-depleted animals. Representative image of a maximum projection of 30-40 sections. n = 10-20 embryos by triplicate. (E) The transmembrane Pom121 nucleoporin was depleted during Zebrafish development and animals were analyzed at 48 hpf. No muscle alterations are observed in the absence of Pom121. Representative image of a maximum projection of 30-40 sections. n = 10-20 embryos by triplicate. (F) Due to the absence of antibodies that recognize Zebrafish Pom121 we verified the effectiveness of Pom121 morpholinos by determining their ability to down-regulate the expression of a fusion RNA containing Pom121 5’UTR upstream of the RFP coding sequence as described by Veldman et al. Pom121 morpholinos efficiently block the translation of the fluorescent reporter (n = 6-10 embryos by duplicate). PHENOTYPE:
|
Reprinted from Developmental Cell, 41, Raices, M., Bukata, L., Sakuma, S., Borlido, J., Hernandez, L.S., Hart, D.O., D'Angelo, M.A., Nuclear Pores Regulate Muscle Development and Maintenance by Assembling a Localized Mef2C Complex, 540-554.e7, Copyright (2017) with permission from Elsevier. Full text @ Dev. Cell