- Title
-
The Hyaloid Vasculature Facilitates Basement Membrane Breakdown During Choroid Fissure Closure in the Zebrafish Eye
- Authors
- James, A., Lee, C., Williams, A.M., Angileri, K., Lathrop, K.L., Gross, J.M.
- Source
- Full text @ Dev. Biol.
|
Temporal and spatial dynamics of basement membrane breakdown during choroid fissure closure in zebrafish. (A) Schematic depicting the approximate level of sections in B-M along the proximal-distal axis of the CF. The vitreous cavity was defined as central, and sections were taken at 12 µm intervals proximally and distally from this point. (B-M) Sagittal sections along the proximal-distal axis of the retina, immunostained for Lam-111 expression. (B-E) 31 hpf, (F-I) 34 hpf, (J-M) 36 hpf. Insets in D, H show high magnification views of the regions in the dashed boxes. (N) Schematic depicting the plane of section for 48 hpf embryos in O,P. (O,P) Representative sagittal sections along the proximal-distal axis of the retina immunostained for Lam-111. Scale bars=20 µm. |
|
in vivo imaging of choroid fissure closure in zebrafish. (A) Schematic depicting the approximate level of sections in B-D along the proximal-distal axis of the CF. The vitreous cavity was defined as central, and optical sections were taken at 16 µm intervals proximally and distally from this point. (B-D) membrane-GFP injected embryos were imaged throughout the CF. Single micron optical slices are shown from 44 to 49 hpf and at three distinct proximal-distal regions of the CF. (B) Distally, the CF remains open until at least 49 hpf. (C) Distal/centrally, the CF appears to close between 46–47 hpf. (D) Proximally, the CF already appears to be closed at 44 hpf. Orange arrowheads in B,C mark open CF. White arrow in C marks what appears to be a closed CF. Dashed line outlines the RPE. Scale bar=50 µm. |
|
Temporal and spatial dynamics of tissue fusion during choroid fissure closure in zebrafish. Single micron optical sections from sagittal cryosections stained with phalloidin (green) and anti-β-catenin (red) at distinct proximal-distal regions of the CF over time. As in Fig. 2, the vitreous cavity was defined as central, and sections were taken at 16 µm intervals proximally and distally from this point. (A-E) At 44 hpf, the CF is fused in central/proximal sections (white arrow) based on co-localization between F-actin and β-catenin in a fusion ‘seam’. (F-J) At 45 hpf, the two sides of the CF are tightly apposed but no fusion outside of the central-proximal region is detected. (K-O) At 47 hpf, a fusion seam is present within central and proximal sections, and there are punctate regions of co-localization in distal/central sections. (P-T) At 49 hpf, the fusion seam has disappeared in the central and proximal CF regions while it is appearing in the distal/central region. (U) Single 1 µm optical sections from one section plane aligned distal (left) to proximal (right) demonstrate a progressive co-localization of F-actin and β-catenin in the CF and formation of the fusion seam. Scale bar=50 µm (A-T) and 5 µm (U). |
|
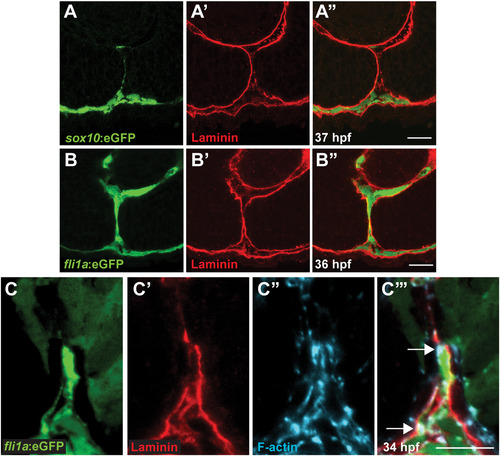
Periocular mesenchymal cells contribute to CFC. (A-C) Sagittal views of the CF stained with anti-GFP (green), Lam-111 (red) and/or phalloidin (blue). (A) Few sox10:eGFP+ cells are detected in the CF, 37 hpf section pictured. (B) fli1a:eGFP+ cells are retained in the CF. 36 hpf section pictured. (C) fli1a:eGFP+ cells possess F-actin accumulations that localize to regions of BM breakdown. Arrows denote puncta of F-actin where Lam-111 is low or absent. 34 hpf section pictured. Scale bar=20 µm (A,B) and 10 µm (C). |
|
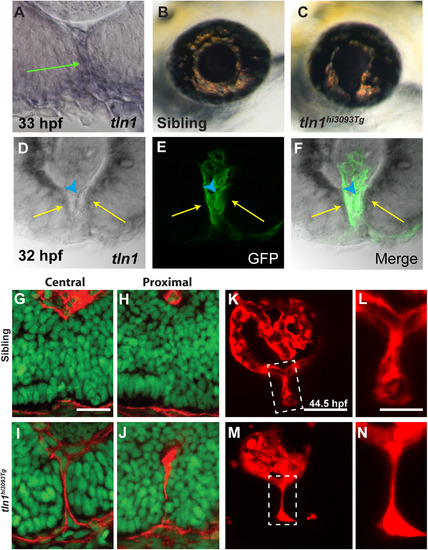
talin1 is required for CFC in zebrafish. (A) talin1 is expressed within the POM and retinal/RPE cells lining the CF at 33 hpf (arrow). (B,C) Lateral views of the eye of tln1hi3093Tg mutant (C) and wild-type sibling (B) at 3dpf. tln1 mutants possess colobomas. (d-F) Distal section through the eye of a 32 hpf embryo demonstrating talin1 expression within the retina/RPE cells lining the CF (arrows) and the hyaloid vasculature (marked by GFP expression from fli1a:eGFP; arrowhead). (G-J) Sagittal sections through the eyes of 48 hpf tln1 mutants and siblings stained with Lam-111 (red) and Sytox-green (DNA; green). BM degradation is disrupted in tln1 mutants at 48 hpf. (K-N) Maximum projection images of the distal hyaloid in tln1 mutants and siblings demonstrating severe hypotrophy of the hyaloid in the tln1 mutant at 44.5 hpf. Scale bar=20 µm (G-J, K,M) and 50 µm (L,N). PHENOTYPE:
|
|
POM-derived endothelial cells facilitate BM breakdown during CFC. (A-D) Sagittal sections through the eyes of 51 hpf clochem378 mutants and siblings stained with Lam-111 (red) and Sytox-green (DNA; green). BM degradation is disrupted in clochem378 mutants in both the (C) proximal and (D) distal regions of the CF when compared to (A,B) siblings. Scale bar=25 µm. PHENOTYPE:
|
|
Periocular mesenchyme cells migrate through the CF during CFC. (A-C) Sagittal section views of the CF immunostained with GFP and Lam-111 antibodies to visualize POM cells (green) and BM (red). Section planes as depicted in Fig. 1A. (A) sox10:eGFP cells at 37 hpf. Few sox10:eGFP cells are detected in the CF. (B,C) fli1a:eGFP cells at 36 hpf at a central and central/proximal plane within the eye. fli1a:eGFP cells spend extended durations in the CF. (D,E) Sagittal section views at central/proximal and distal/central planes of the eye showing fli1a:eGFP cells (green) and sox10:memRFP cells (red) in the CF at 34 hpf. Scale bar =20 µm. |

Unillustrated author statements PHENOTYPE:
|
Reprinted from Developmental Biology, 419(2), James, A., Lee, C., Williams, A.M., Angileri, K., Lathrop, K.L., Gross, J.M., The Hyaloid Vasculature Facilitates Basement Membrane Breakdown During Choroid Fissure Closure in the Zebrafish Eye, 262-272, Copyright (2016) with permission from Elsevier. Full text @ Dev. Biol.