- Title
-
Minor class splicing shapes the zebrafish transcriptome during development
- Authors
- Markmiller, S., Cloonan, N., Lardelli, R.M., Doggett, K., Keightley, M.C., Boglev, Y., Trotter, A.J., Ng, A.Y., Wilkins, S.J., Verkade, H., Ober, E.A., Field, H.A., Grimmond, S.M., Lieschke, G.J., Stainier, D.Y., and Heath, J.K.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
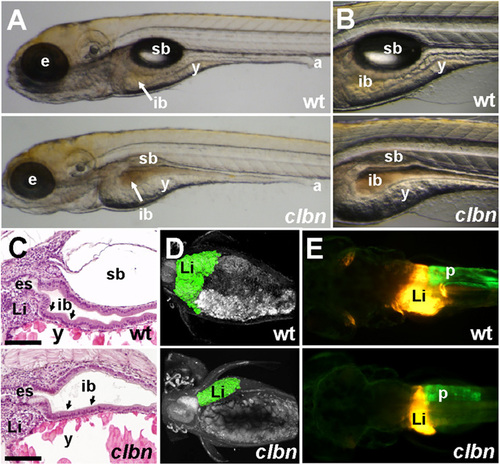
The clbn phenotype is characterized by abnormalities in the digestive organs (A and B) Brightfield images of left lateral views and (C) histology of WT and clbns846 larvae at 120 hpf show a thick, folded intestinal epithelium (black arrows in C) and inflated swim bladder in WT, whereas clbn displays a thin, unfolded intestinal epithelium, delayed yolk resorption, and smaller eyes. (D) Two-photon microscopy of WT and clbns846 larvae on the Tg(gutGFP)s854 background and (E) epifluorescence microscopy on the Tg(fabp10:RFP,ela3l:EGFP)gz12 background reveal smaller digestive organs in clbns846 compared with WT. The two images in A were taken at the same magnification, as were the two images in each of panels B, D, and E. The clbn phenotype is 100% penetrant and all larvae die between 7 and 10 dpf; however, the severity of the morphological abnormalities may vary between clutches. For example, the digestive organ phenotype is visibly conspicuous in some clutches at around 108 hpf and not in others until 120 hpf. a, anus; e, eye; es, esophagus; ib, intestinal bulb; Li, liver; p, pancreas; sb, swim bladder; y, yolk. (Scale bar in C, 100 µm.) |
|
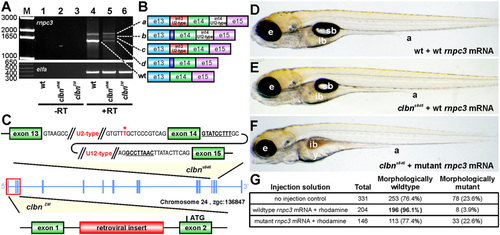
The clbn phenotype is caused by a mutation in rnpc3. (A) RT-PCR of full-length rnpc3 coding sequence amplifies a predominant 1.6-kb product in WT larvae (lane 4) and four aberrantly spliced transcripts in clbns846 (lane 5). A retroviral insertion in clbnZM abolishes generation of full-length rnpc3 mRNA (lane 6). Lanes 1?3 show ?RT controls. (Lower) Elongation factor alpha (elfa) control. Dashed lines link the cDNA products in lanes 4 and 5 to the corresponding transcripts, shown schematically in B. The dark blue region denotes retention of 10 intronic nucleotides through use of a de novo 32 ss. (C) Two noncomplementing clbn alleles. An asterisk over T marks the position of the T′A variation, 12 nucleotides upstream of exon 14 of rnpc3, which causes the clbns846 phenotype. The conserved 52 ss and BPS in intron 14 (U12-type) of rnpc3 are underlined and in bold. clbnZM carries a retroviral insertion in the first intron of rnpc3. (D?F) Brightfield microscopy images of left lateral views of 5-dpf larvae derived from a clbns846 heterozygous incross that were injected at the one- to two-cell stage with 380 pg of either WT rnpc3 mRNA or a mutant mRNA retaining introns 13 and 14, corresponding to the most abundant form of rnpc3 mRNA in clbns846 (see B). (D) No significant overexpression phenotype in WT larvae injected with WT rnpc3 mRNA. (E) Rescue of the mutant phenotype in genotyped clbns846 homozygous mutant larvae injected with WT rnpc3 mRNA. (F) No rescue of the clbn mutant phenotype injected with mutant rnpc3 mRNA. (G) Noninjected control larvae from the same clutch show a near-perfect Mendelian ratio of WT to clbn larvae, whereas 96% of larvae injected with WT rnpc3 RNA appear WT at 5 dpf (P < 0.0001, Fisher?s exact test). There was no difference between uninjected and mutant rnpc3-injected groups (P = 0.9065). a, anus; e, eye; ib, intestinal bulb; sb, swim bladder; y, yolk. The brightfield images in D, E, and F were taken at the same magnification. |
|
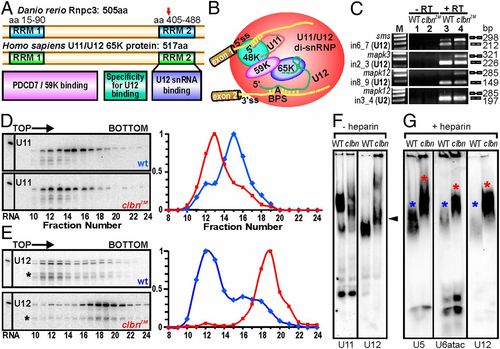
Zebrafish Rnpc3 functions in the U12-type spliceosome. (A) Domain structure of zebrafish Rnpc3 and its human ortholog RNPC3, or U11/U12 di-snRNP 65-kDa protein (www.uniprot.org, accession no. Q96LT9). Red arrow in the C-terminal RRM denotes the approximate position of the premature stop codons in all clbns846 isoforms. (B) Schematic of the human U11/U12 di-snRNP, with RNPC3 (65 K) bridging U12 snRNA and the 59 K protein (28). BPS, branch point sequence; ?A? represents the branch point adenosine. (C) RT-PCR analysis of three different transcripts (sms, mapk3, mapk12) shows retention of U12-type introns (lane 4, Top three panels) in clbns846 (5 dpf), but not of a U2-type intron (Bottom). Lanes 1 and 2 show ?RT controls. (D and E) Glycerol gradient/Northern analysis revealing differential sedimentation of U11 (D) and U12 (E) snRNA-containing snRNPs in WT and clbnZM extracts. Direction of sedimentation is left to right. Lane 1 contains total RNA. Full-length U11 or U12 signals were quantified using ImageQuant and expressed as a percentage of the fraction with the highest intensity. U12 snRNA showed persistent specific degradation (asterisks) without affecting its migration profile. (F) Northern analysis of WT and clbns846 extracts resolved on 4% (80:1) native polyacrylamide gels and probed for U11 and U12 snRNAs. The predominant U11 snRNPs are disrupted in clbn compared with WT and the U12-containing particles are heavier and migrate more slowly (black arrowhead). (G) Northern analysis of WT and clbns846 extracts resolved on native gels and probed for U5, U6atac, and U12 snRNAs shows retarded bands in clbn (red asterisks) compared with WT (blue asterisks). The same larval lysate was used in F and G. Data are representative of a total of 10 (D and E) and 5 (F and G) biological replicates. PHENOTYPE:
|
|
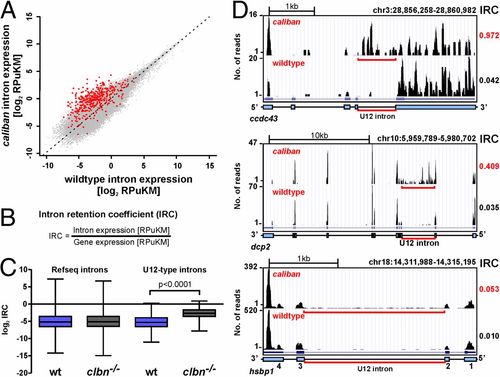
Intron retention in clbn is specific to U12-type introns and highly variable. (A) Scatterplot of normalized intron expression derived by RNAseq for reference (Refseq) introns (gray dots) and U12-type introns (red dots). U12-type introns lie almost entirely above the midline representing equal expression, indicating enhanced intron retention in clbn. Intron expression is given in RPuKM. (B) To quantitate U12-type intron retention in individual transcripts, we defined an intron retention coefficient, which calculates intron retention relative to transcript level. (C) Comparison of the median log2 of the IRCs for Refseq introns and U12-type introns between WT and clbn demonstrates that the clbn splicing defect is specific to U12-type introns. See also Fig. S4. Statistical significance was determined by Student t test. (D) University of California at Santa Cruz genome browser views of three genes with high, intermediate, and low IRCs. ccdc43 shows a high IRC of 0.97, dcp2 an intermediate IRC of 0.41, and hsbp1 shows a low IRC of 0.05. PHENOTYPE:
|
|
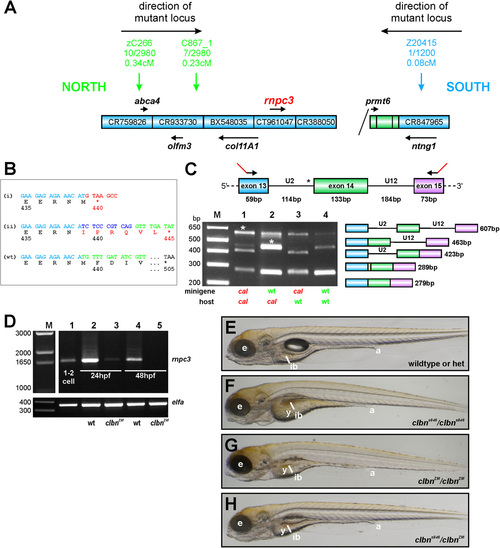
rnpc3 is the mutated locus in clbn. (A) Simple schematic representation of the genetic interval containing the mutated locus in clbns486 based on the zebrafish genome assembly (Zv9). The genetic region of interest was defined by determining linkage of several simple sequence length polymorphism (SSLP) markers to the mutation (1). A total of up to 2,980 meioses from a defined series of crosses between the original ethylnitrosourea-mutagenized strain and a polymorphic mapping strain were analyzed, narrowing down the genetic interval containing the mutation to a distance of 0.31 cM between the closest linked markers. Blue boxes represent bacterial artificial chromosome (BAC) clones encompassing the region. Diagonal line indicates the presence of additional intervening sequence. Black arrows and gene names shown in italics represent annotated genes. (B) Aberrant transcripts extracted from clbns846 larvae encode truncated translation products. RT-PCR of rnpc3 RNA extracted from clbns846 larvae produces four faint bands (see Fig. 2A), corresponding to the aberrant transcripts (a?d) shown schematically in Fig. 2B. Product (i) is encoded by transcripts retaining intron 13 (a and c in Fig. 2B). A frame-shift in the coding sequence results in a premature stop codon instead of residue 440. Product (ii) is encoded by transcripts (b and d in Fig. 2B) using a de novo 32 splice site (ss) in intron 13 that results in the addition of 10 intronic nucleotides to the coding sequence. This also introduces a frame-shift and a premature stop codon is encountered instead of residue 445. Full-length WT product is shown below. (C) In vivo minigene assay to characterize the splicing defect in clbns846. A minigene spanning the U2-type intron 13, the U12-type intron 14, and flanking exons, was amplified from WT and clbns846 genomic DNA using primers carrying 52 tails (23 bp; shown in red) to allow specific amplification of the exogenous transgene after injection into zebrafish embryos. No correctly spliced RT-PCR product is detected upon injection of mutant minigene plasmid DNA into clbns846 and WT embryos (lanes 1 and 3, respectively). Sequencing of the lower bands confirms the use of the same de novo 32 ss detected endogenously in clbns846. The retention of 10 intronic nucleotides is indicated by the yellow region in the schematic diagram. Mutant larvae display significant retention of the U12-type intron 14 upon injection of both the WT and mutant minigene (lanes 1 and 2, asterisks). (D) Full-length rnpc3 cDNA is RT-PCR amplified from RNA extracted from one to two cell stage embryos (lane 1) indicating maternal deposits of WT rnpc3. The level of full-length rnpc3 is greatly reduced in clbnZM mutants by 24 hpf and undetectable by 48 hpf. Lanes contain RT-PCR products of RNA extracted from genotyped homozygous WT (lanes 2 and 4) and clbnZM mutant embryos (lanes 3 and 5) at 24 hpf (lanes 2 and 3) and 48 hpf (lanes 4 and 5). (Lower) elfa control. (E?H) Brightfield microscopy images of left lateral views of (E) WT siblings from a clbnZM × clbns846 cross, (F) homozygous mutant clbns846 larvae, and (G) homozygous mutant clbnZM larvae. (H) Twenty-five percent of offspring from clbnZM × clbns846 crosses show a phenotype indistinguishable from that of either clbns846 or clbnZM homozygous mutants. The images in panels E?H were taken at the same magnification. PHENOTYPE:
|
|
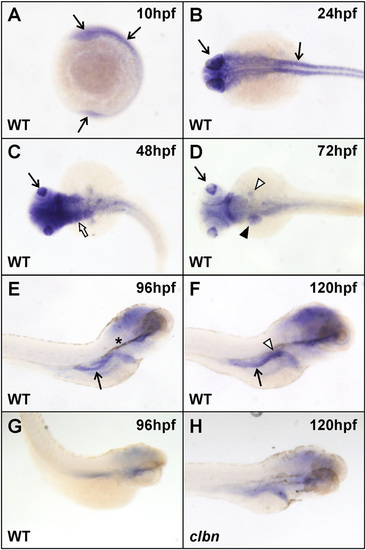
Widespread rnpc3 mRNA expression in early zebrafish embryos becomes more restricted during development. WISH analysis was carried out on clutches of embryos derived from an in-cross of clbnZM heterozygotes using a 599-bp antisense RNA probe designed to hybridize to zebrafish rnpc3 mRNA. (A and B) Widespread rnpc3 mRNA expression in WT embryos at 10 and 24 hpf (arrows). (C and D) From 48 to 72 hpf, rnpc3 mRNA expression becomes progressively restricted to proliferating tissues, including the lens (arrow), pharyngeal region (white arrow), liver and pancreas anlage (black and white arrowheads, respectively), and intestine. (E and F) At 96?120 hpf, rnpc3 mRNA expression is prominent in the digestive organs. Right lateral views show strong expression in the developing pancreas (white arrowhead) and intestine (arrow). (G) Sense probe at 96 hpf produces very weak, nonspecific staining. (H) At 120 hpf, clbn larvae show markedly reduced rnpc3 expression, compared with WT (F). B?D are dorsal views; E?H are right lateral views. Asterisk in E denotes residual pigment after PTU treatment. The same patterns of expression were obtained with a 790-bp probe designed to hybridize to a nonoverlapping region of rnpc3 mRNA. All embryos/larvae shown were genotyped and all images were taken at the same magnification. |