Fig. 4
- ID
- ZDB-FIG-140806-2
- Publication
- Markmiller et al., 2014 - Minor class splicing shapes the zebrafish transcriptome during development
- Other Figures
- All Figure Page
- Back to All Figure Page
|
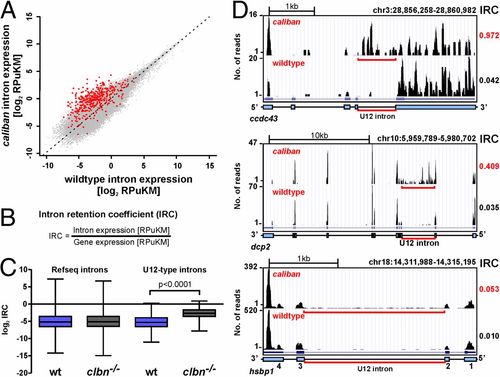
Intron retention in clbn is specific to U12-type introns and highly variable. (A) Scatterplot of normalized intron expression derived by RNAseq for reference (Refseq) introns (gray dots) and U12-type introns (red dots). U12-type introns lie almost entirely above the midline representing equal expression, indicating enhanced intron retention in clbn. Intron expression is given in RPuKM. (B) To quantitate U12-type intron retention in individual transcripts, we defined an intron retention coefficient, which calculates intron retention relative to transcript level. (C) Comparison of the median log2 of the IRCs for Refseq introns and U12-type introns between WT and clbn demonstrates that the clbn splicing defect is specific to U12-type introns. See also Fig. S4. Statistical significance was determined by Student t test. (D) University of California at Santa Cruz genome browser views of three genes with high, intermediate, and low IRCs. ccdc43 shows a high IRC of 0.97, dcp2 an intermediate IRC of 0.41, and hsbp1 shows a low IRC of 0.05. |
| Fish: | |
|---|---|
| Observed In: | |
| Stage: | Day 4 |