Fig 1
- ID
- ZDB-FIG-200229-20
- Publication
- Concilio et al., 2020 - Inter-species variation in monovalent anion substrate selectivity and inhibitor sensitivity in the sodium iodide symporter (NIS)
- Other Figures
- All Figure Page
- Back to All Figure Page
|
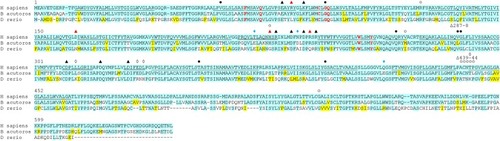
Cyan highlighting indicates absolute conservation to human NIS. Yellow indicates similar residue to human NIS. Underline indicates putative transmembrane domain in human NIS, only TM1-12 are indicated. Closed circles indicate site of a mutation known to cause a transport defect in humans [ |