Fig. 1
- ID
- ZDB-FIG-160816-1
- Publication
- Venero Galanternik et al., 2015 - Heparan Sulfate Proteoglycans Regulate Fgf Signaling and Cell Polarity during Collective Cell Migration
- Other Figures
- All Figure Page
- Back to All Figure Page
|
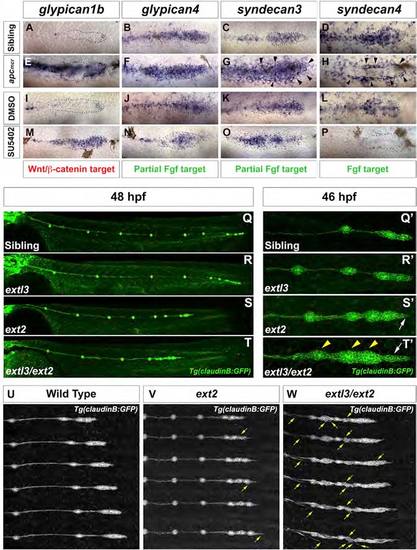
Loss of HSPGs Results in LL Prim Migration and Patterning Defects (A, E, I, and M) gpc1b expression in (A) WT, (E) apcmcr, (I) DMSO-treated, and (M) SU5402-treated embryos. (B, C, F, G, J, K, N, and O) gpc4 and sdc3 expression in (B and C) WT, (F and G) apcmcr, (J and K) DMSO-treated, and (N and O) SU5402-treated embryos. (D, H, L, and P) sdc4 expression in (D) WT, (H) apcmcr, (L) DMSO-treated, and (P) SU5402-treated embryos. (G and H) Black arrowheads point to the ectopic halo of sdc3 and sdc4. (Q and R) WT and extl3 mutant prim has reached the tail tip by 48 hpf. (S and T) ext2 and extl3/ext2 prim fails to complete migration. (Q′-T′) Magnification of prim at 46 hpf. (Q′ and R′) WT and extl3 mutant prim shows a normal morphology. (S′) ext2 mutant prim possesses a pointed tip (white arrow). (T′) extl3/ext2 mutant prim shows severe morphological defects. The leading edge elongates (white arrow) and rosettes are closely spaced (yellow arrowheads). (U-W) Still images of time-lapse movies: WT (U), ext2 (V), and extl3/ext2 mutant prim (W). Yellow arrows in (V) and (W) point to ectopic filopodia. See also Figure S2 and Movies S1, S2, S3, and S4. |
| Genes: | |
|---|---|
| Fish: | |
| Condition: | |
| Anatomical Terms: | |
| Stage Range: | Prim-15 to Long-pec |
| Fish: | |
|---|---|
| Condition: | |
| Observed In: | |
| Stage Range: | Prim-15 to Long-pec |