Fig. 7
|
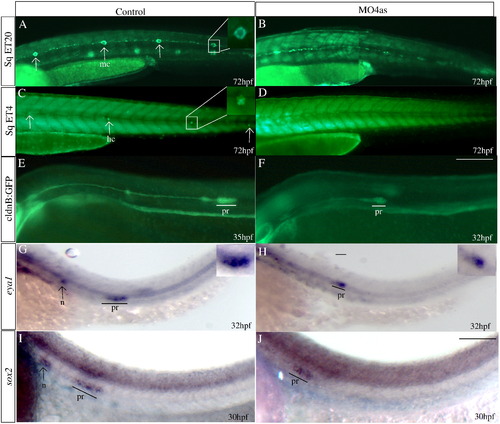
irx4a is required for the correct development of the posterior lateral line. GFP expression was observed in the SqET20 (A, B), SqET4 (C, D) and cldnB:GFP (E, F) transgenic lines in control and morphant larvae at the indicated times post-fertilization. In situ hybridization was carried out using the eya1 (G, H) and sox2 (I, J) riboprobes. (A, B) Expression of GFP in 72 hpf control SqET20 transgenic line labels mantle cells (arrows in A, inset) and interneuromastic cells (inc). Morphant embryos (B) lack neuromast mantle cells but interneuromastic cells are present indicating that primordium migration occurred. (C, D) At 72 hpf mechanosensory hair cells (arrows in C, inset) are labeled in the SqET4 transgenic line while morpholino injected siblings (D) show the absence of these cells. (E, F) The migrating primordium, the trail of interneuromastic cells and deposited neuromasts express GFP in cldnB:GFP transgenic uninjected embryos (E). In contrast, MO4as injected cldnB:GFP embryos (F) show a decrease in the size of the migrating primordium and the absence of deposited neuromasts. Note that we have compared GFP label in a 32 hpf morphant with a 35 hpf control embryo. The primordium size difference is evident despite the fact that the primordium in the control fish has already deposited two neuromasts. (G?J) Expression of the primordium markers eya1 (G, H, insets show magnified views) and sox2 (I, J) shows a similar effect, the label is diminished in morphant embryos. inc, interneuromastic cells hc, hair cells; n, neuromast; pr, primordium. The entire panel shows lateral views of the trunk and tail region; dorsal is towards top, anterior is towards left. Scale bar in F: 50 μm for A?F, scale bar in J: 30 μm for G?J. |
| Genes: | |
|---|---|
| Fish: | |
| Knockdown Reagent: | |
| Anatomical Terms: | |
| Stage Range: | Prim-15 to Protruding-mouth |
Reprinted from Molecular and cellular neurosciences, 40(3), Feijoo, C.G., Saldias, M.P., De la Paz, J.F., Gómez-Skarmeta, J.L., and Allende, M.L., Formation of posterior cranial placode derivatives requires the Iroquois transcription factor irx4a, 328-337, Copyright (2009) with permission from Elsevier. Full text @ Mol. Cell Neurosci.