- Title
-
Effects of realgar-indigo naturalis formula on a zebrafish tumor xenograft model induced by human acute promyelocytic leukemia cells: antitumor activity, hepatotoxicity, and transcriptomic analysis
- Authors
- Bai, D., Zhang, Z., Gao, J., Wang, Q., Macdonald, R., Xu, Z., Chen, S., Huang, N., Luo, L.
- Source
- Full text @ Front Pharmacol
|
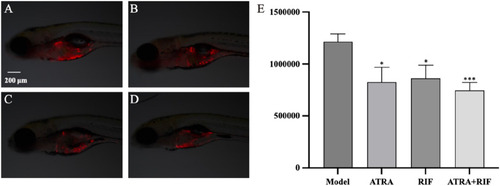
Growth of zebrafish tumor cells (HL - 60) after sample treatment |
|
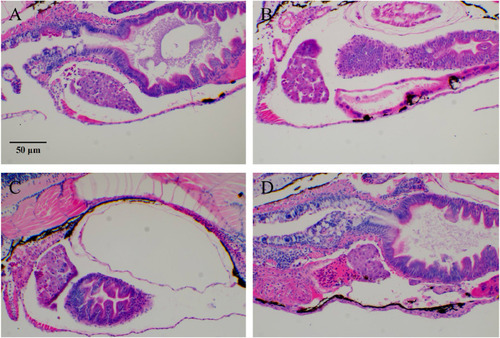
Typical images of zebrafish tumor cell (HL-60) migration after RIF intervention. |
|
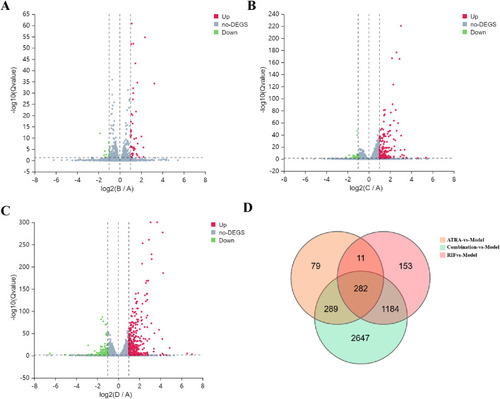
Transcriptomic profiling of differentially expressed genes (DEGs) across treatment groups. |
|
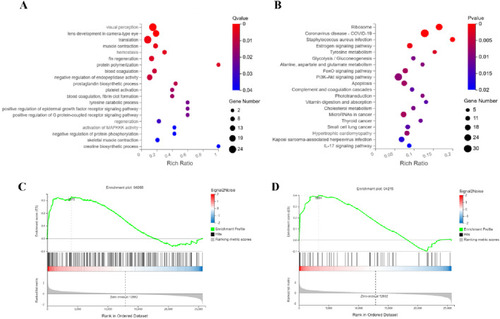
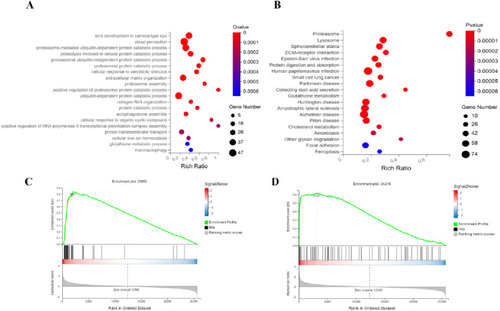
Integrated mechanisms of ATRA in APL therapy. Functional analyses identify key pathways mediating ATRA’s therapeutic effects: |
|
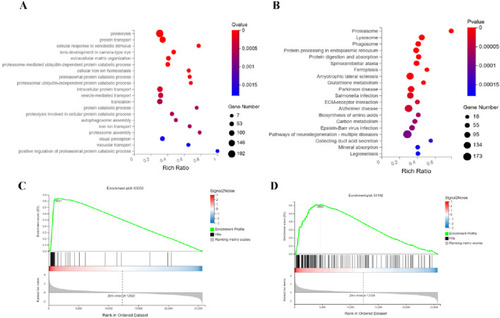
Synergistic targeting of proteasome and ferroptosis pathways underlies RIF’s anti-leukemic efficacy. Functional dissection identifies RIF-driven pathway perturbations: |
|
Mechanism of the combination therapy in synergistically suppressing APL through triple-axis targeting of the proteasome, ferroptosis, and lysosome pathways. |