- Title
-
Cancer-associated fibroblast driven paracrine IL-6/STAT3 signaling promotes migration and dissemination in invasive lobular carcinoma
- Authors
- Bullock, E., Rozyczko, A., Shabbir, S., Tsoupi, I., Young, A.I.J., Travnickova, J., Gómez-Cuadrado, L., Mabruk, Z., Carrasco, G., Morrow, E., Pennel, K., Kloosterman, P., Houthuijzen, J.M., Jonkers, J., Avalle, L., Poli, V., Iggo, R., Xiao, X., Guo, J., Zhu, X., Mallon, E., Edwards, J., Patton, E.E., Brunton, V.G.
- Source
- Full text @ Breast Cancer Res.
|
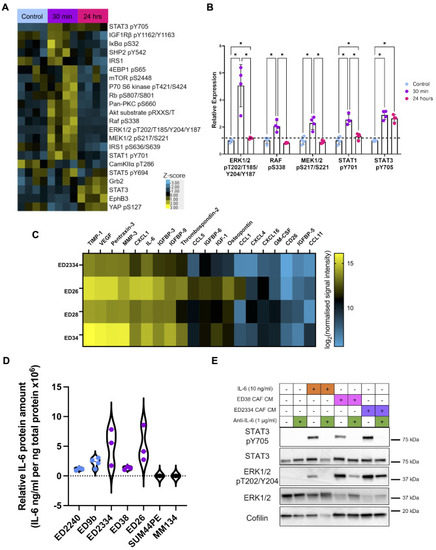
ILC CAF conditioned media drives STAT3 and MAPK pathway activation. ( |
|
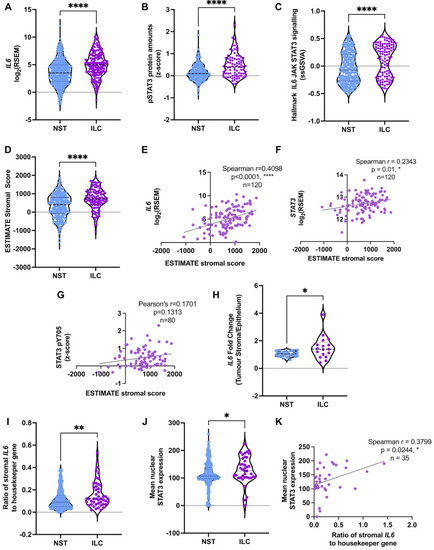
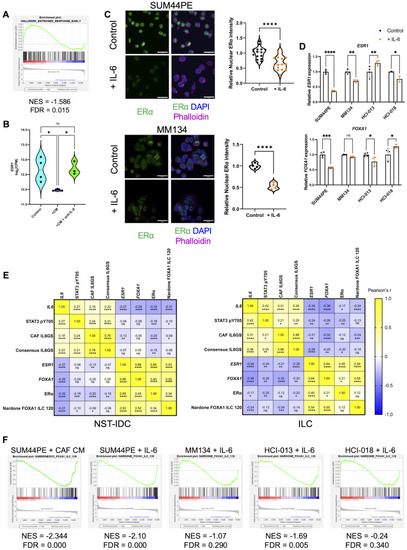
IL-6/STAT3 pathway is augmented in ER + ILC compared to ER + NST tumors. ( |
|
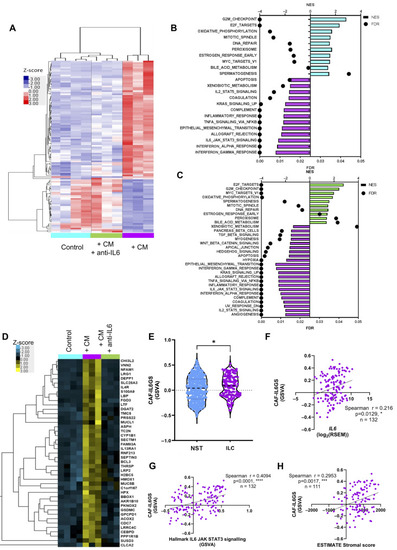
IL-6 in CAF conditioned media drives gene expression in ILC cells. ( |
|
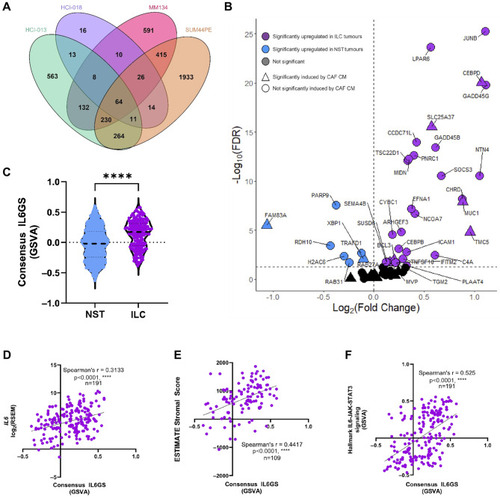
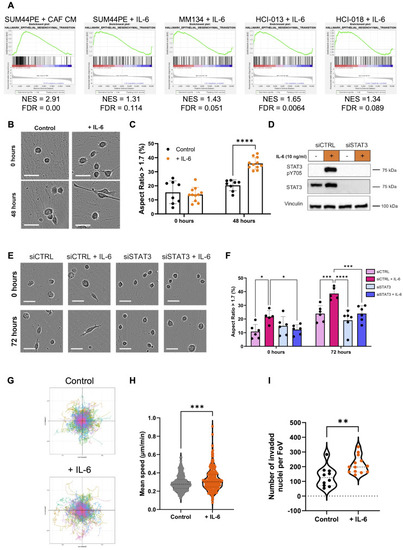
IL-6 drives consistent gene changes in multiple ILC models. ( |
|
IL-6 regulates estrogen receptor and FOXA1 expression in ILC tumor cells. ( |
|
IL-6 drives a mesenchymal and pro-migratory phenotype in SUM44PE cells. ( |
|
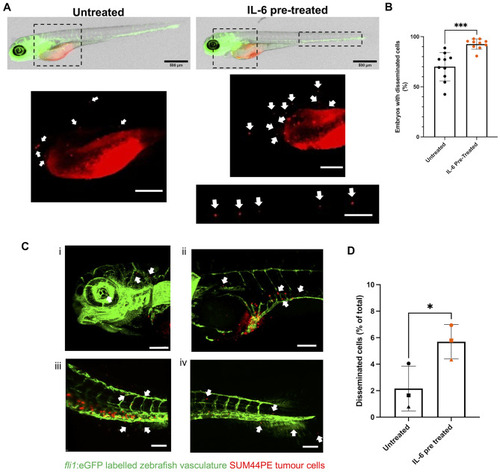
IL-6 promotes increased dissemination of SUM44PE cells in zebrafish embryos. ( |