- Title
-
Impact of HOMER2 frameshift extension variant on auditory function and development
- Authors
- Han, E., Kim, J.A., Park, S., Han, J.H., Kim, M.Y., Kim, Y., Tran, N.T., Kim, B.J., Choi, J., Choi, B.Y.
- Source
- Full text @ J. Mol. Med.
|
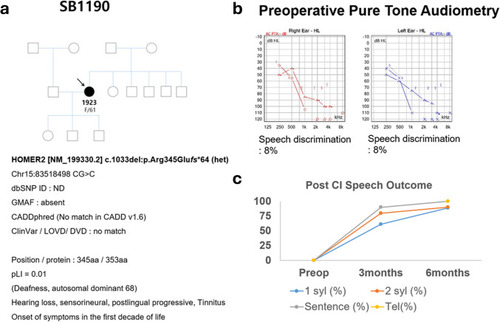
Pedigree, audiometric profile, genetic analysis, and post-cochlear implantation speech outcomes from SB1190-1923 (F/61). |
|
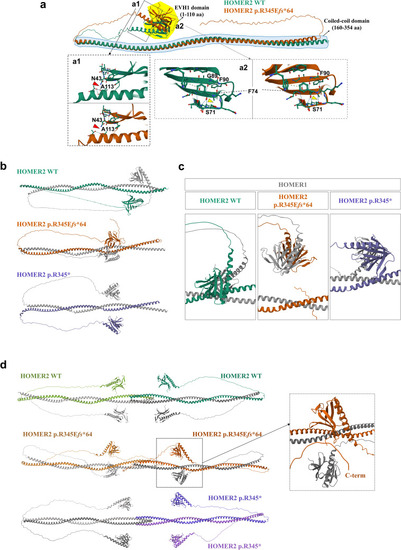
Predicted 3D structures of HOMER2 WT and HOMER2 p.R345E |
|
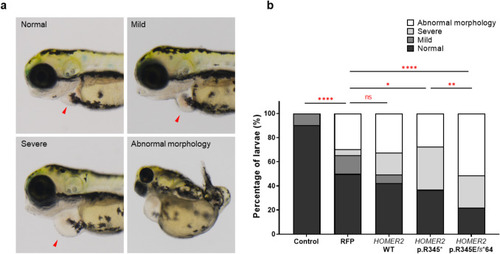
Cardiac and morphological defects in zebrafish larvae injected with |
|
Otic capsule area in zebrafish larvae at 3 days post-fertilization. |
|
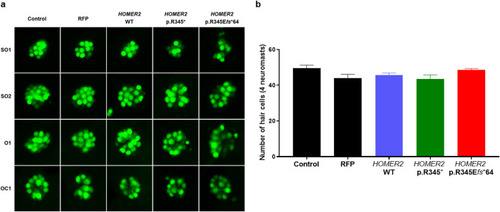
Analysis of hair cell numbers in zebrafish neuromasts. |
|
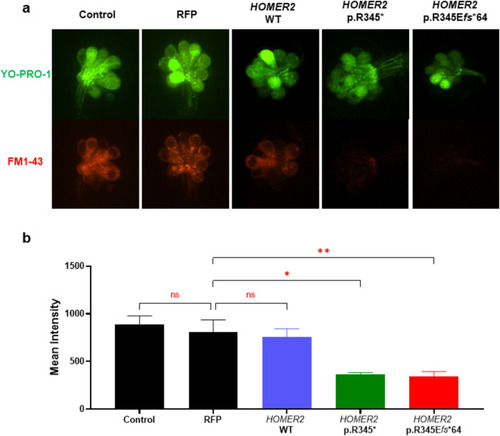
Comparison of FM1-43 uptake in zebrafish neuromast hair cells. |
|
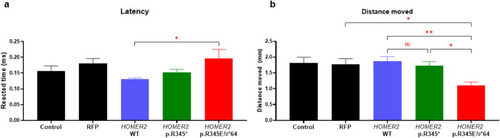
Comparison of startle reflex in zebrafish larvae at 6 days post-fertilization. |