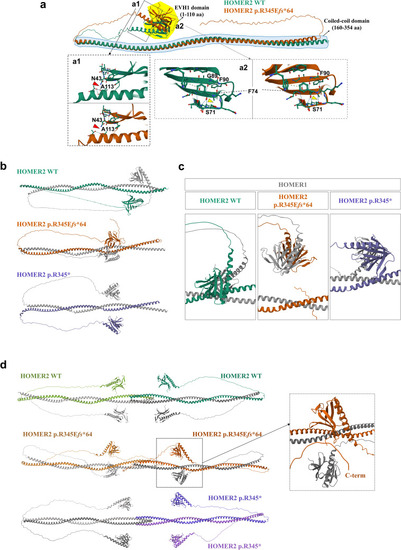
Predicted 3D structures of HOMER2 WT and HOMER2 p.R345Efs*64 using AlphaFold2. a Structural alignment of HOMER2 WT and HOMER2 p.R345Efs*64. The EVH1 domain is highlighted in the yellow area, and the coiled-coil (CC) domain is marked by the blue dashed line. a1 The hydrogen bond between N43 and A113 is disrupted in HOMER2 p.R345Efs*64 compared to HOMER2 WT. a2 The β-sheet structure in HOMER2 p.R345Efs*64 extends to F90, and a hydrogen bond forms between F74 and S71, which is absent in HOMER2 WT. b The structure of the HOMER1 [NP_004263.1] and HOMER2 dimer is shown. HOMER1 is depicted in gray, while HOMER2 WT, HOMER2 p.R345Efs*64, and HOMER2 p.R345* are represented in green, brown, and purple, respectively. The coiled-coil region of HOMER2, which can interact with other proteins, is located at amino acids 307–329 in the WT and is shifted to amino acids 275–297 in the HOMER2 variants. c Structural changes in the EVH1 domain were detected in the HOMER1 WT and HOMER2 variants dimer. The distance between each EVH1 domain of HOMER1 and HOMER2 p.R345Efs*64 was reduced (b) compared to the WT, which is shifted vertically. In HOMER1 WT and HOMER2 p.R345* dimer, the distance between each EVH1 was increased (b) and slightly shifted vertically. d The predicted tetramer structures were obtained using HOMER1 and HOMER2. Two HOMER1 proteins were combined with two HOMER2 WT, two HOMER2 p.R345Efs*64, or two HOMER2 p.R345* molecules. In the HOMER1 and HOMER2 p.R345Efs*64 tetramer, the location of the EVH1 domain of HOMER1 was altered, and the C-terminal of HOMER2 approached the EVH1 domain of HOMER2 (dashed black box). In the HOMER1 and HOMER2 p.R345* tetramer, HOMER1 and HOMER2 formed homodimers, and each homodimer formed a tetramer, unlike the other configurations. WT, wild-type; R, arginine; E, glutamate; fs, frameshift; EVH1, Ena/Vasp homology domain 1; F, phenylalanine; S, serine; N, asparagine; A, alanine
|

