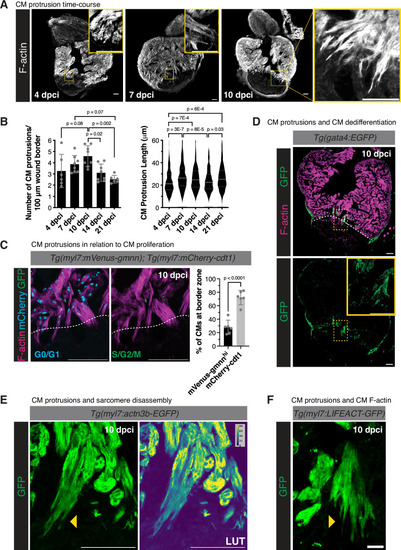
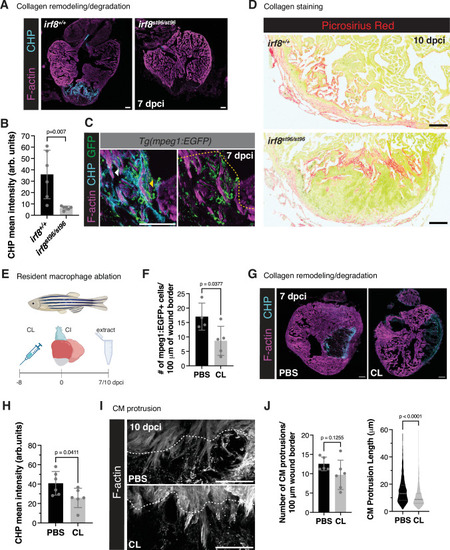
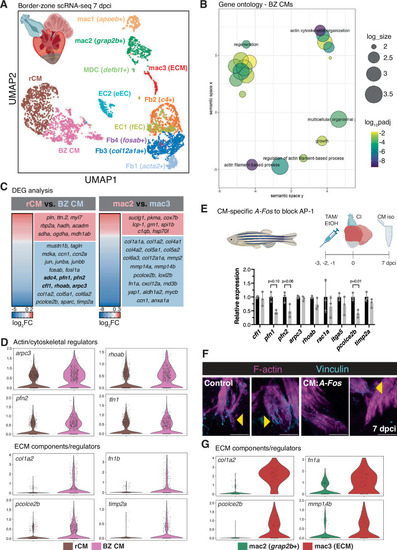
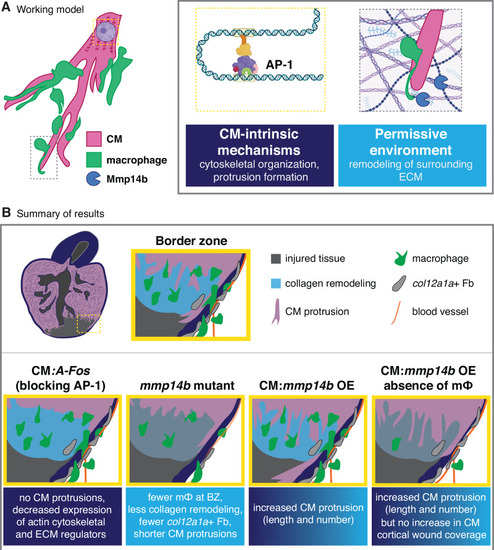
|
Mmp14b is a regulator of CM protrusion.A Collagen hybridizing peptide (CHP) and phalloidin staining of a wild-type ventricle at 10 dpci. Yellow box denotes the area in the zoomed image. B In situ hybridization of mmp14b expression in a wild-type ventricle at 7 dpci. Black dashed line denotes the approximate injury border. C Colocalization of mmp14b (HCR, magenta) with CMs (MHC immunostaining), endocardial cells (vwf HCR), macrophages (GFP immunostaining in Tg(mpeg1:EGFP) ventricles, and fibroblasts (col12a1a (HCR, green) and postnb (HCR, cyan) in the cortical BZ region at 10 dpci. Yellow box in the schematic of the heart marks the cortical CM region depicted in the zoomed images. Created in BioRender. Beisaw, A. (2025) https://BioRender.com/zek72tx. D Schematic depicting the exon structure of the mmp14b locus and CRISPR/Cas9-induced full-length deletion between exons 2 and 9 of mmp14b. Created in BioRender. Beisaw, A. (2025) https://BioRender.com/pepiu5b. E Mmp14b wild-type and putative mutant protein domain structure (left). RT-PCR of the mmp14b open reading frame from wild-type and mmp14bΔ/Δ mutant embryos (right). SP signal peptide, Pro propeptide, Cat catalytic domain, H hinge region, TM transmembrane domain, C C-terminal tail. F RT-qPCR of mmp14b and mmp14a expression in single ventricles from mmp14bΔ/Δ (n = 5 ventricles) and wild-type siblings (n = 5 ventricles) at 10 dpci. Data are presented as mean ± SD. P-values were calculated using an unpaired two-sided t-test. Source data are presented in the Source Data file. G Phalloidin staining of thick cryosections from mmp14bΔ/Δ and wild-type sibling ventricles at 10 dpci (left). Quantification of CM protrusion length (right, mmp14bΔ/Δn = 1124 CM protrusions from 8 ventricles, wild-type sibling n = 1480 CM protrusions from 9 ventricles). Data are presented as violin plots of all points with solid gray lines indicating the median and dotted gray lines indicating 25th and 75th percentile. P-values were calculated using a two-sided Mann–Whitney test. Source data are presented in the Source Data file. H Picrosirius red staining of collagen in mmp14bΔ/Δ (n = 8 ventricles) and wild-type sibling (n = 10 ventricles) at 60 dpci (left). Quantification of scar area (% of ventricle area) on the right. Data are presented as mean ± SD. P-value was calculated using an unpaired two-sided t-test. Source data are presented in the Source Data file. I Quantification of CM proliferation within 100 μm of the wound border from PCNA/Mef2 immunostaining in mmp14bΔ/Δ (n = 3 ventricles) and wild-type sibling (n = 4 ventricles) at 7 dpci. Data are presented as mean ± SD. P-value was calculated using an unpaired two-sided t-test. Source data are presented in the Source Data file. Scale bars: 100 μm in (A, B, G, and H), 20 μm in zoomed image in (A and C).
|