- Title
-
RACK1 contributes to the upregulation of embryonic genes in a model of cardiac hypertrophy
- Authors
- Ceci, M., Bonvissuto, D., Papetti, F., Silvestri, F., Sette, C., Catalani, E., Cervia, D., Gornati, R., Romano, N.
- Source
- Full text @ Sci. Rep.
|
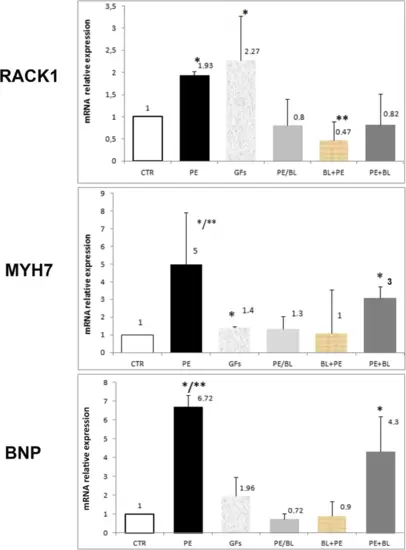
Quantitative RT‒PCR analysis of RACK1 and hypertrophy markers. The graph shows the relative expression of Rack1 (RACK1), myosin heavy chain 7 (MYH7), and brain natriuretic peptide (BNP). In the PE-treated samples, a significant increase in markers was observed compared with those in the control samples (PE vs. CTR; PE vs. all groups p < 0.001,*). The GFs group, despite the variability of the samples, was significantly greater than the CTR and the other groups were, except for the PE group (GFs vs. CTR, PE/BL, PE + BL BL + PE, *, p < 0.01). The PE/BL and PE + BL groups presented nonsignificant modifications, whereas the BL + PE group presented a reduction (BL + PE vs. CTR, **, p < 0,05). MYH7 expression was significantly upregulated in the PE and PE + BL groups (PE vs. CTR, *, PE vs. all groups, **, p < 0.001; PE + BL vs. all groups without PE, *, p < 0.005). Compared with the CTR group, the GFs group presented a slight increase (GFs vs. CTR, *, p < 0.05). BNP was increased in the PE group (PE vs. CTR; p < 0.001; *; PE vs. all other groups). **, p < 0.05) and the PE + BL group (PE + BL vs. CTR, *; p < 0.01). Conversely, this upregulation was partially repressed after pre- and cotreatment with BL. The GFs also presented a greater mean than did the control, but the variability among the samples was not statistically significant. Hearts from the fish in each group (n = 6) were subjected to triplicate experiments—statistical analysis: ANOVA with the Bonferroni correction. |
|
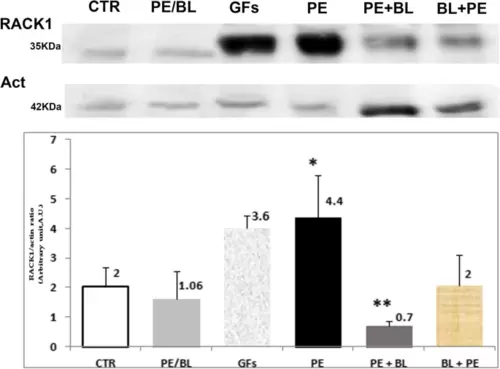
Immunoblot analysis of RACK1 protein expression in the experimental groups vs. CTR. Immunoblot images of Rack1 protein (RACK1) expression (upper part) and nonsarcomere actin (below) were used as housekeeping proteins. The graph shows the means of at least three independent experiments of the density ratio between the RACK1 and actin bands (mean ± S.D.). Compared with that in the CTR group, RAC1 expression was strongly marked in the PE group (p < 0,001, *), but appreciable positivity was also observed in the GFs in the CTR group (P < 0,05, *). The level of RACK1 was comparable to that of CTR in the other treatment groups. The PE + BL group expressed less protein than the CTR group did (**; p < 0.05). Hearts from the fish used in triplicate experiments (n = 10 in each group) were subjected to statistical analysis: ANOVA with the Bonferroni correction. |
|
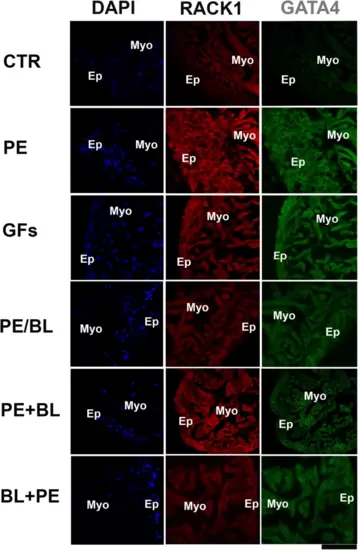
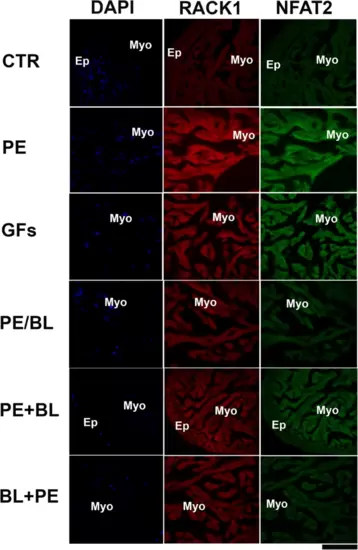
Double staining with RACK1 and GATA4 was analysed via confocal microscopy. RACK1 (red fluorescence) is fundamentally expressed in the CTR myocardium. In all the experimental groups, RACK1 was increased and colocalized with the GATA4 antibody (green fluorescence). GATA4 is strongly expressed in PE and GFs and moderately expressed in PE + BL and PE/BL. Moreover, it was expressed at lower levels in the BL + PE treatment. The hearts utilized for the experiments were N = 4/5 sections in each group in triplicate experiments. Blue fluorescence: DAPI (nuclear marker); scale bar: 500 μm. |
|
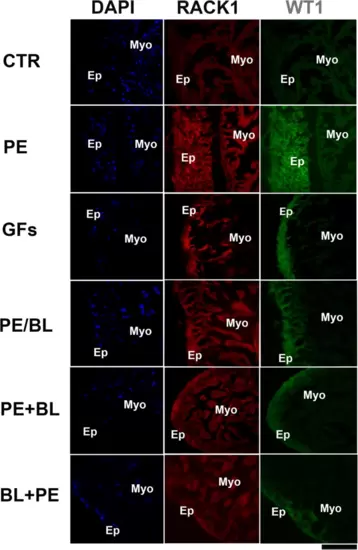
Double staining of RACK1 and WT1 was performed via confocal microscopy. RACK1 (red fluorescence) is expressed at lower levels in the epicardium than in the myocardium in the CTR. In all the experimental groups, RACK1 expression was increased, and RACK1 was particularly colocalized with the WT1 antibody, which stains the epicardium (green fluorescence). WT1 is strongly expressed in PE and GFs and moderately expressed in PE + BL, PE/BL, and BL + PE. The hearts utilized for the experiments were N = 4/5 sections in each group in triplicate experiments. Blue fluorescence: DAPI (nuclear marker); scale bar: 500 μm. |
|
Double staining of RACK1 and NFAT2 was performed via confocal microscopy. RACK1 (red fluorescence) is fundamentally expressed in the myocardium of CTR. In all the experimental groups, RACK1 expression was increased, and RACK1 colocalized with the NFAT2 antibody to mark the endocardium (green fluorescence). NFAT2 is strongly expressed in PE and GFs and moderately expressed in PE + BL, PE/BL, and BL + PE. The hearts utilized for the experiments were N = 4/5 sections in each group in triplicate experiments. Blue fluorescence: DAPI (nuclear marker); scale bar: 500 μm. |
|
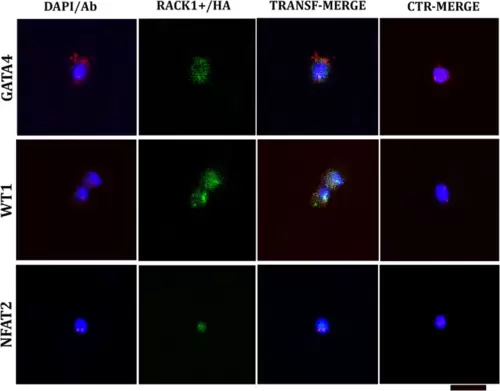
Confocal analysis at 24 h post-transfection (T0). The plasmid containing rack1+ and the reporter hemagglutinin (RACK1+/HA) produced green fluorescence. The cells were double-labelled with embryonal markers (GATA4, WT1, and NFAT2; red fluorescence) and compared to nontransfected cells (CTR). GATA4 is increased in transfected cells and is localized mainly in the cytoplasm. WT1 is comparable to the CTR. NFAT2 is sparsely expressed in the CTR and, in both cases, is localized in the nucleus. The nucleus was labelled with DAPI (blue fluorescence). DAPI/Ab: DAPI-labelled antibody (GATA4, WT1 or NFAT2). TRANSF/MERGE: Transfected cells labelled with all the antibodies (against HA and one embryonal marker). CTR/MERGE: nontransfected cells labelled with all the antibodies. The hearts utilized for the experiments were N = 4/5 sections in each group in triplicate experiments. The hearts utilized for the experiments were N = 3, and analysis was performed with 104 cells in each group in triplicate. Bar: 20 μm. |
|
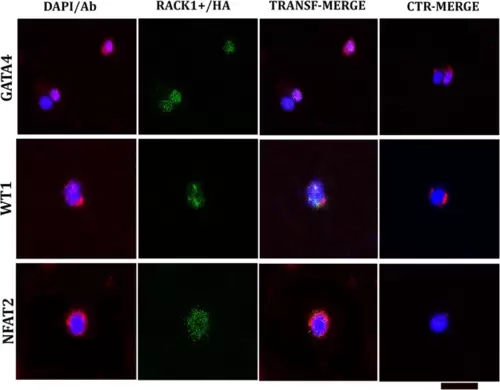
Confocal microscopy analysis after transfection and one week of culture in medium supplemented with GFs (T1). The plasmid containing RACK1/HA emitted green fluorescence. The cells were double-labelled with embryonal markers (GATA4, WT1, and NFAT2; red fluorescence) and compared to nontransfected cells (CTR). GATA4 is strongly increased in transfected cells and is localized mainly in the nucleus. Compared with CTR, WT1 is highly expressed in the nucleus and cytoplasm. Compared with its expression in the nucleus, NFAT2 is more highly expressed in the cytoplasm than CTR, which is localized in the nucleus. Nuclei are marked with DAPI (blue fluorescence); DAPI/Ab: DAPI labelling + antibody (GATA4 or WT1 or NFAT2). TRANSF/MERGE: transfected cells labelled with all the antibodies (against HA and one embryonal marker). CTR/MERGE: nontransfected cells labelled with all the antibodies. The hearts utilized for the experiments were N = 3, and analysis was performed with 104 cells in each group in triplicate. Bar: 20 μm. |
|
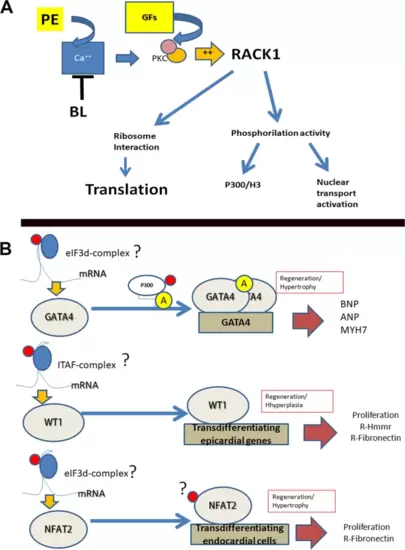
RACK1 activation (A) and hypotheses about the possible direct interaction of RACK1 with embryonal marker expression (B). A) According to previous studies, RACK1 can be activated by PKC, which, in turn, is activated by calcium waves induced by PE or a cocktail of FGFs (GFs). BL treatment depletes the calcium wave. B) The translation of the mRNA of GATA4 could involve the IRES sequence because our bioinformatic analysis revealed a long sequence in the 5’ UTR. RACK1 interacts with the ITAF/IRES complex to permit ribosome assembly. The acetylation of GATA4 by the transcriptional coactivator p300 induces its multimerization and activates its DNA binding activity. GATA4 permits the transcription of cardiac hypertrophic response genes, including atrial natriuretic factor (ANF), brain natriuretic peptide (BNP), and myosin heavy chain 7 (MYH7). Preliminary bioinformatic analysis revealed that the long 5’UTR of WT1 is compatible with the IRES sequence. RACK1 can interact with the ITAF/IRES complex for ribosome assembly during translation. WT1, as a transcription factor, can induce epicardial cell proliferation and transdifferentiation. NFAT2 can interact with RACK1 in two ways: translation via the eIF-3d 5’UTR and cooperation in transporting the phosphorylated protein to the nucleus. NFAT2 is a powerful transcription factor for the proliferation and transdifferentiation of the endocardium and endothelial cells. |