- Title
-
Ciliogenesis defects after neurulation impact brain development and neuronal activity in larval zebrafish
- Authors
- D'Gama, P.P., Jeong, I., Nygård, A.M., Trinh, A.T., Yaksi, E., Jurisch-Yaksi, N.
- Source
- Full text @ iScience
|
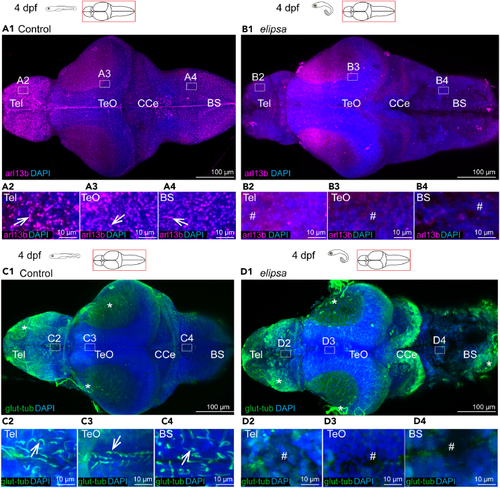
Loss of primary and motile cilia in the (A1–A4 and B1–B4) Staining of dissected 4 dpf brains with arl13b antibody to stain all cilia in the brain, (B1) In the (C1) At 4 dpf, single glutamylated tubulin-positive cilia were present in the forebrain choroid plexus (C2), on the dorsal roof and ventral part (C3) of the tectal/diencephalic ventricle and in the rhombencephalon choroid plexus (C4). (D1) In See also |
|
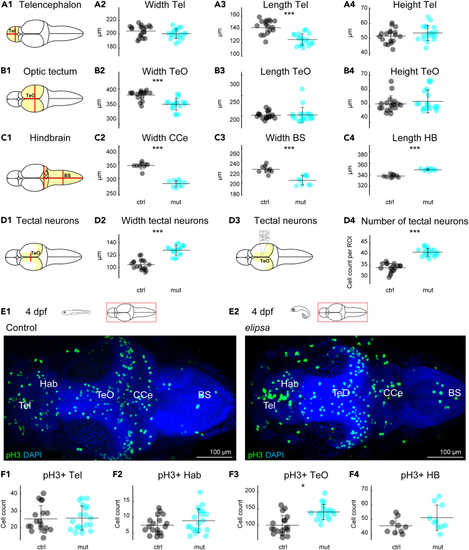
Cilia defects lead to abnormal brain size and alters cell proliferation in the optic tectum (A1–D4) Quantification of brain morphology of 4 dpf larval brains for control (black) and (E1 and E2) staining for mitotic cells using an anti-pH3 antibody. (F1–F4) Cell count for pH3 positive cells (pH3+) in telencephalon (F1), habenula (F2), optic tectum (F3) and hindbrain (F4). ∗: |
|
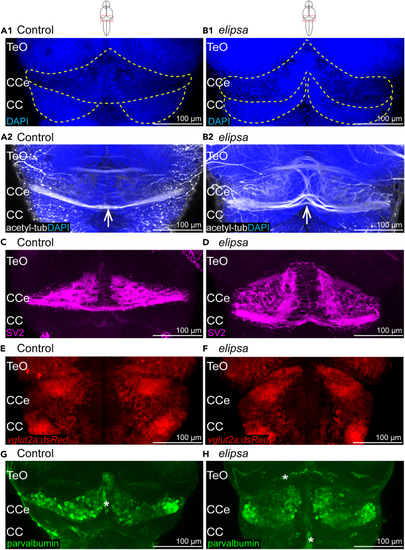
Cerebellar defects in 4 days old (A1–B1) Cell nucleus stained using dapi in control (A1) and (C and D) Larvae immunostained with the presynaptic vesicle marker SV2 in control (C) and |
|
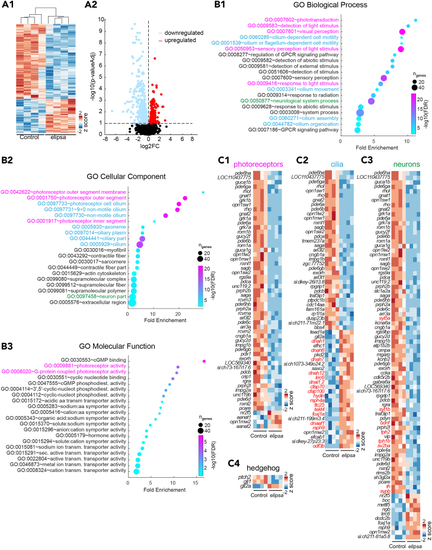
Transcriptomic analysis reveals altered expression of genes involved in phototransduction (A1) Heatmap of expression of differentially expressed genes (DEGs) from control and (A2) Volcano plot showing the DEGs in (B) Top 20 Gene Ontology (GO) biological process (B1) cellular component (B2) and molecular function (B3). Genes associated with GO terms related to phototransduction (magenta), cilia (blue) and neuronal process (green) are represented in (C1–C3). (C) Heatmaps showing expression of DEGs (represented as See also |
|
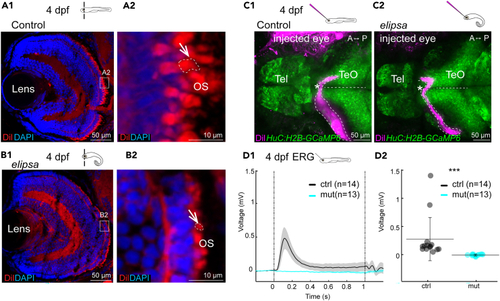
(A1–B2) DiI staining of 4 dpf retina cryosection to stain the outer segments. Section through the whole retina or the photoreceptor layer of a representative control (A1-A2) and (D1-D2) Electroretinography (ERG) recordings in a 4 dpf retina. D1 Average response of electrical activity (+/− standard error of the mean as the shaded region) to 1 s light stimulation for control (black) and mutant (cyan). D2 Average electrical responses for the 200 msec following the light ON stimulus for all control fish (black) and |
|
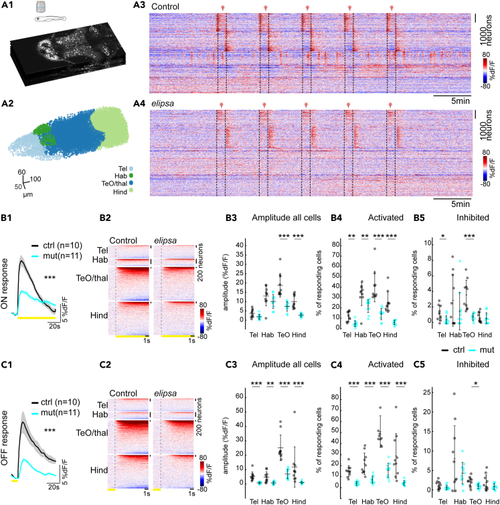
Reduced photic-induced neural activity in the brain of (A1) Optical sections of multiplane recording of a transgenic zebrafish larva expressing nuclear GcaMP6s in all neurons (B1, C1) Neural responses to light stimulation for control (black) and mutant (cyan) averaged over 5 trials of photic stimulations for ON (B1) and OFF (C1) response conditions (+/− standard error of mean as the shaded region trace). (B2, C2) Activity of neurons per brain regions during ON (B2) and OFF response (C2) for representative examples (warm color represents higher calcium signals). (B3, C3) Average amplitude of all cells during ON (B3) and OFF (C3) response for control (black) and mutant (cyan), amplitude was significantly reduced in TeO and Hind regions for ON response and all brain regions for OFF response. (B4, C4 and B5, C5) % of cells that are activated during ON (B4) and OFF response (C4) and inhibited during ON (B5) and OFF response (C5). ∗: PHENOTYPE:
|
|
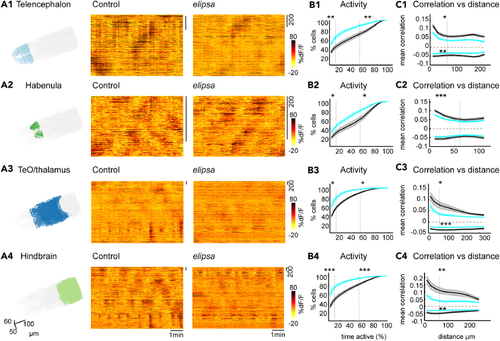
Reduced ongoing spontaneous activity and correlation in the (A1–A4) Three-dimensional representation of segmented neurons and their activity for one representative control and (B1–B4) Average cumulative frequency distribution graphs showing the percentage of activity of neurons in Telencephalon (B1), Habenula (B2), TeO/thalamus (B3) and Hindbrain (B4), for control (black) and (C1–C4) Mean Pearson’s correlation versus distance graphs showing positive and negative correlation between cells in Telencephalon (C1), Habenula (C2), TeO/thalamus (C3) and Hindbrain (C4), for control (black) and mutant (cyan). Significance tests for the correlation were computed for cells located within 60 μm indicated by the dotted line on the X axis. ∗: See also |