- Title
-
Somatic PDGFRB activating variants promote smooth muscle cell phenotype modulation in intracranial fusiform aneurysm
- Authors
- Hao, L., Ya, X., Wu, J., Tao, C., Ma, R., Zheng, Z., Mou, S., Ling, Y., Yang, Y., Wang, J., Zhang, Y., Lin, Q., Zhao, J.
- Source
- Full text @ J. Biomed. Sci.
|
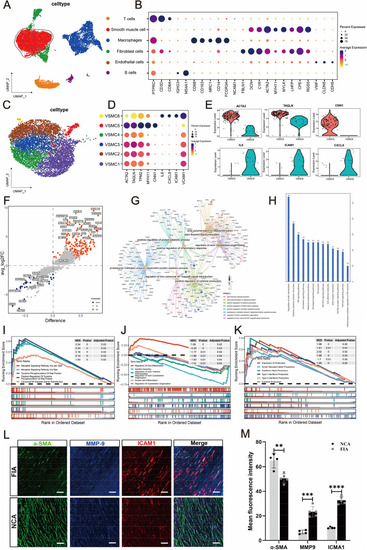
Single-cell transcriptional profiling of intracranial fusiform aneurysmal cells ( |
|
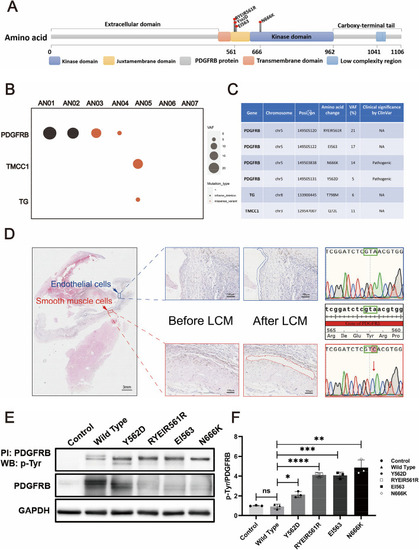
Somatic mutational landscape of FIAs. Structure of the protein encoded by PDGFRβ and the lollipop plots illustrate the location of the identified mutations in our cohort. The color of the boxes denotes different regions of the encoded protein. The coordinates represent the n-th amino acids ( |
|
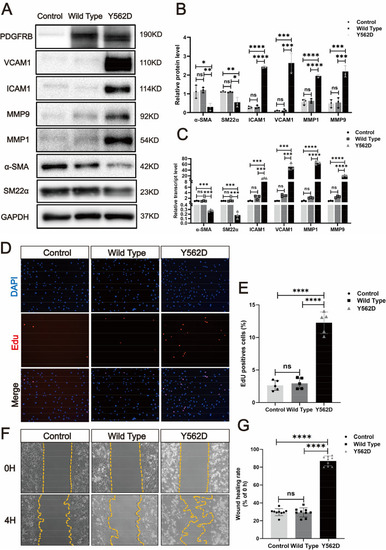
PDGFRB somatic mutation induce phenotypic modulation in SMCs. Immunostaining reveals the expression levels of smooth muscle markers (a-SMA and SM22a) and inflammatory markers (VCAM1, ICAM1, MMP1 and MMP9) in HBVSMCs transfected with different viruses (Control: vector; Wild Type: PDGFRB; Y562D: PDGFRBY562D) ( |
|
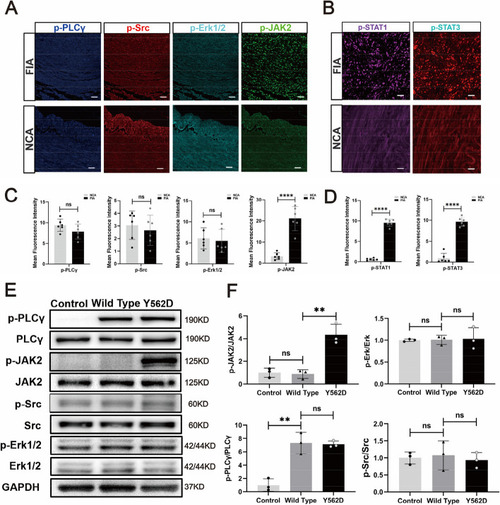
The JAK2-STAT pathway serves as the major downstream signaling pathway of the mutated PDGFB. mIF demonstrate the important downstream signaling pathway markers of PDGFRβ (p-JAK, p-Src, p-Erk1/2 and p-PLCγ) in FIA sections and NCA sections ( |
|
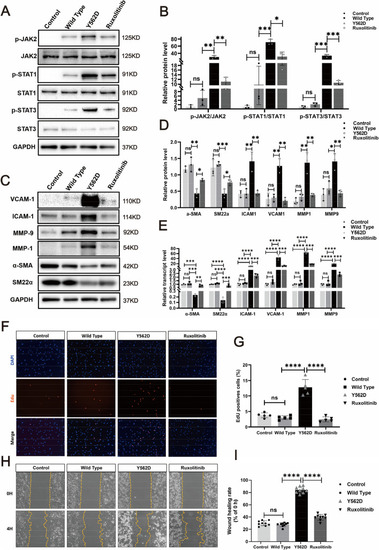
Ruxolitinib effectively reverse the PDGFRBY562D-induced phenotypic modulation in HBVSMCs. Western blotting illustrates the alterations in p-JAK2 and p-STAT (p-STAT1 and p-STAT3) expression across different treatment groups ( |
|
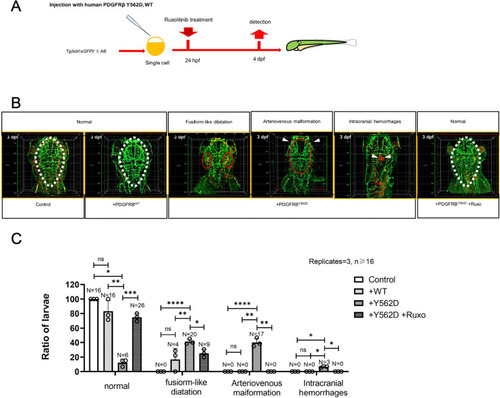
Ruxolitinib can reverse the phenotype caused by the PDGFRβ.Y562D in zebrafish. |