- Title
-
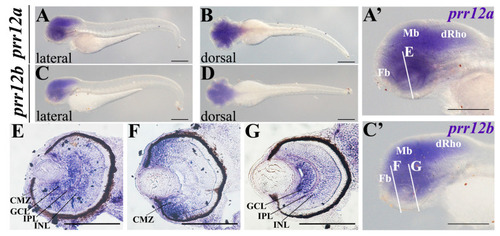
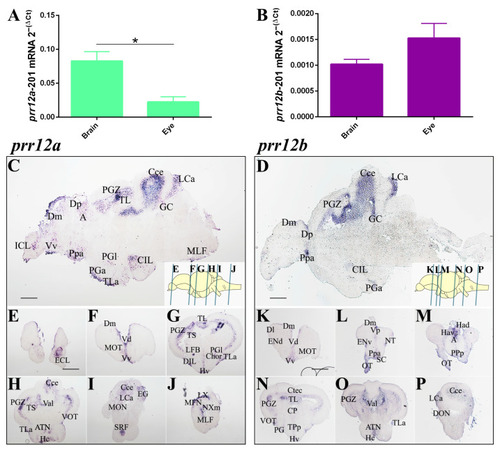
Shedding a Light on Dark Genes: A Comparative Expression Study of PRR12 Orthologues during Zebrafish Development
- Authors
- Muscò, A., Martini, D., Digregorio, M., Broccoli, V., Andreazzoli, M.
- Source
- Full text @ Genes (Basel)
|
|
|
EXPRESSION / LABELING:
|
|
EXPRESSION / LABELING:
|
|
EXPRESSION / LABELING:
|
|
EXPRESSION / LABELING:
|
|
EXPRESSION / LABELING:
|