- Title
-
Transcriptional control of visual neural circuit development by GS homeobox 1
- Authors
- Schmidt, A.R., Placer, H.J., Muhammad, I.M., Shephard, R., Patrick, R.L., Saurborn, T., Horstick, E.J., Bergeron, S.A.
- Source
- Full text @ PLoS Genet.
|
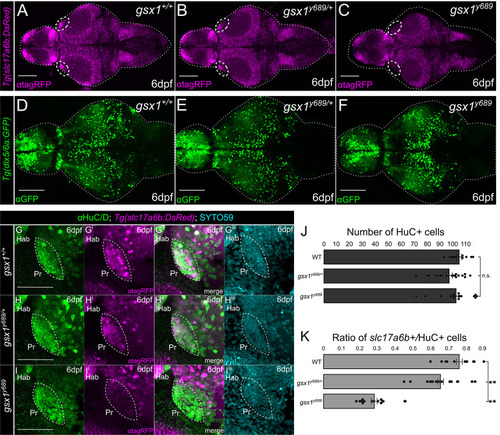
Examining excitatory and inhibitory neuron differentiation in |
|
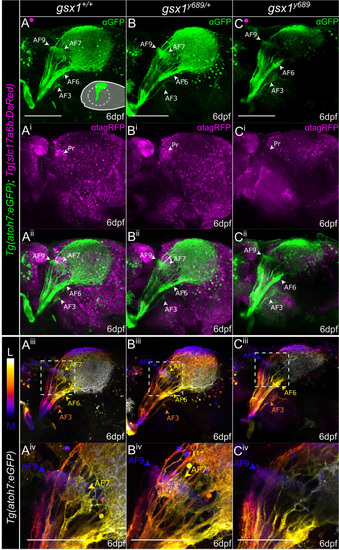
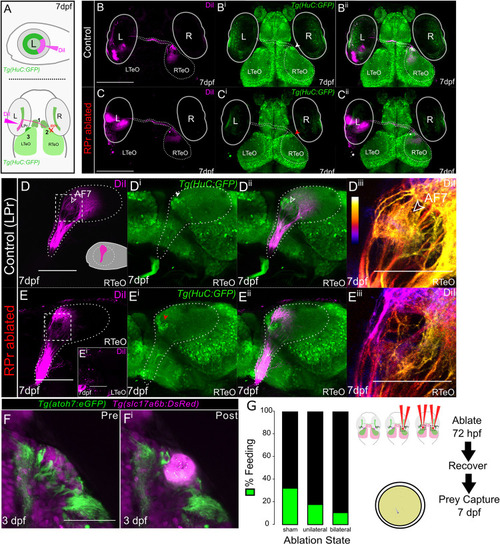
RGC axon termination is disrupted in |
|
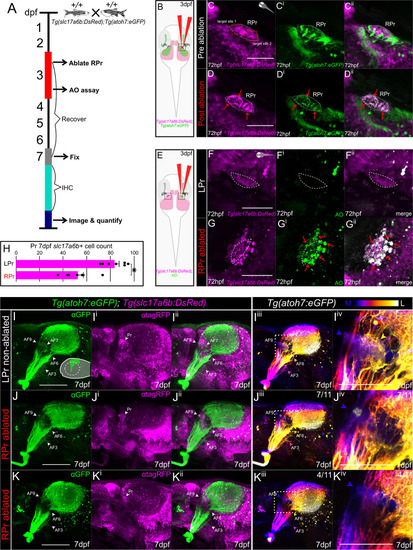
RGC axon volume and trajectory examination in |
|
|
|
|
|
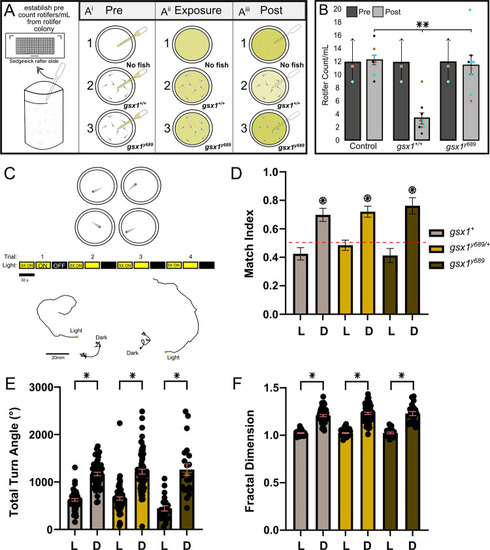
Retinotopographic order and prey capture defect is confirmed after pretectal ablations in the AF7 region. |
|
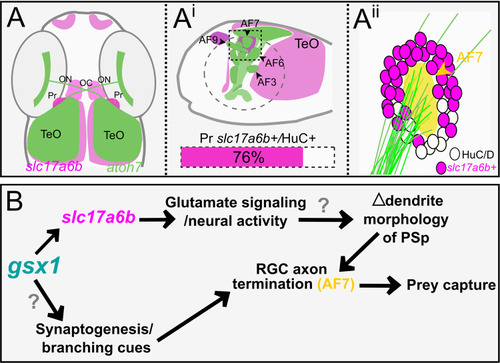
A model for |