- Title
-
Molecular basis of retinal remodeling in a zebrafish model of retinitis pigmentosa
- Authors
- Santhanam, A., Shihabeddin, E., Wei, H., Wu, J., O'Brien, J.
- Source
- Full text @ Cell. Mol. Life Sci.
|
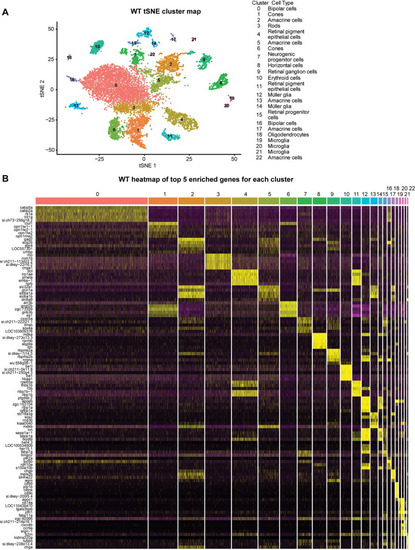
Single-cell RNA seq analysis of WT zebrafish retina. |
|
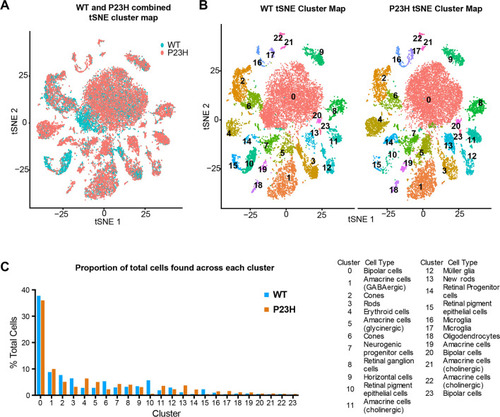
Single-cell RNA seq analysis of integrated WT-P23H zebrafish retina datasets. |
|
Transcriptomic analysis of rods and newly formed rods in the WT-P23H integrated dataset reveal an increased response to oxidative stress and misfolded proteins. |
|
Comparative transcriptomic analysis of cones in WT-P23H integrated dataset reveals changes in metabolism and circadian regulation. |
|
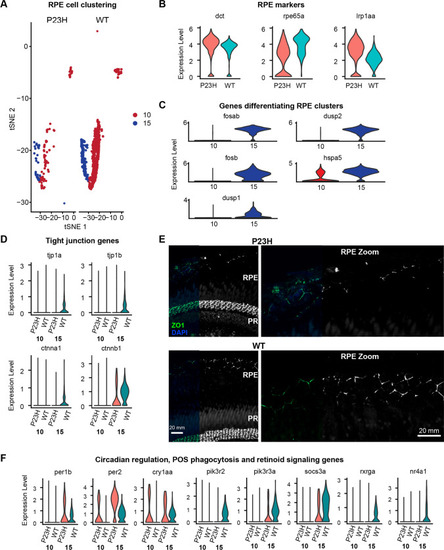
RPE transcriptome is altered in the RP model. |
|
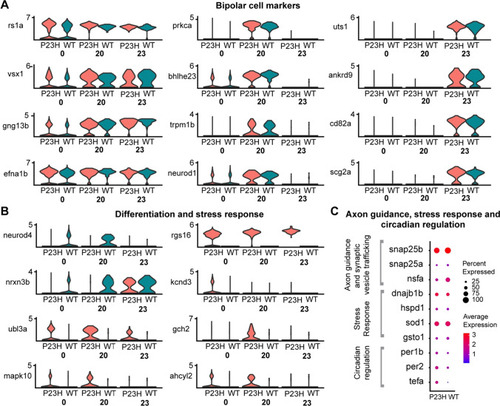
Comparative transcriptomic analysis of bipolar cell clusters in the WT-P23H integrated dataset shows evidence of stress and neuronal remodeling in the RP model. |
|
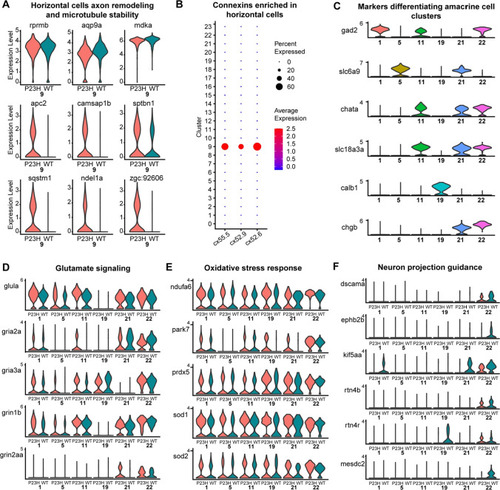
Transcriptomic analysis of Horizontal and Amacrine cells in the WT-P23H integrated dataset reveals changes in axon remodeling in the RP model. |
|
Comparative transcriptomic analysis of retinal ganglion cells in WT-P23H integrated dataset. |
|
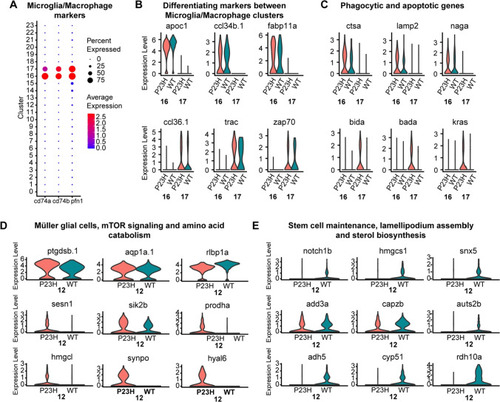
Comparative transcriptomic analysis of microglial/macrophage clusters and Müller glial cells in the zebrafish retina |
|
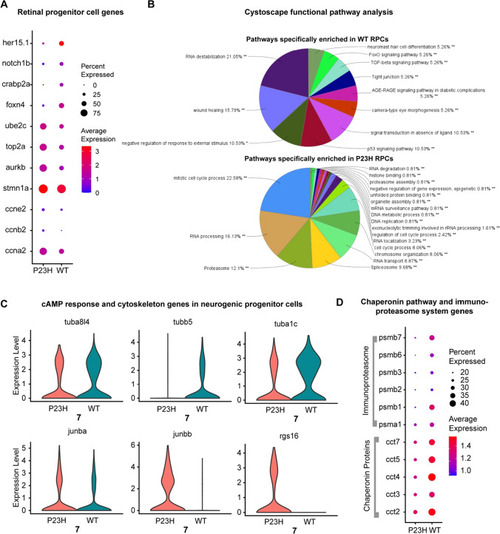
Retinal progenitor cells (RPCs) are enriched in the RP model whereas a neurogenic progenitor cell (NPCs) cluster was identified in both WT and P23H |