- Title
-
Dual Action of Eeyarestatin 24 on Sec-Dependent Protein Secretion and Bacterial DNA
- Authors
- Schäfer, A.B., Steenhuis, M., Jim, K.K., Neef, J., O'Keefe, S., Whitehead, R.C., Swanton, E., Wang, B., Halbedel, S., High, S., van Dijl, J.M., Luirink, J., Wenzel, M.
- Source
- Full text @ ACS Infect Dis
|
Summary of B. subtilis transcriptomic stress profiling results. (A) Chemical structures of ES24 and NFT. The nitrofuran ring is highlighted in red. (B) Number of regulated transcripts per regulon. Sec components and proteases potentially indicating secretion stress are highlighted. NfrAB is a nitro/flavin reductase under transcriptional control of the Spx regulator and among the most strongly activated transcripts for both ES24 and NFT treatment. (C) Number of induced transcripts per functional category of the respective gene products. Functional categories were assigned according to function, regulation, interaction, and expression data available on http://subtiwiki.uni-goettingen.de/. Transcripts that showed a log2-fold change of ≥3 (p < 0.05) were considered as differentially regulated. |
|
Oxidative stress assays. (A) Oxyburst Green assay for detecting ROS in B. subtilis 168CA. The fluorescent probe shows a marked increase in fluorescence when ROS are present. It should be noted that Oxyburst Green has been reported to be specific for the detection of superoxide, yet we have found that it reacts to different sources of ROS (see Figure S2). (B) Influence of ROS scavengers on antibiotic activity against B. subtilis 168CA. MICs were determined in the absence or presence of the superoxide scavenger tiron or the hydroxyl radical scavenger thiourea. If one of these ROS species contributes to the activity of the compound, the MIC should increase in the presence of the respective scavenger. |
|
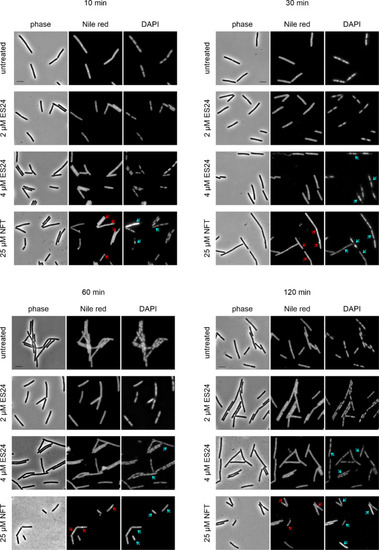
Bacterial cytological profiling of B. subtilis 168CA treated with ES24 or NFT. Red arrows indicate membrane aberrations, and blue arrows indicate nucleoid aberrations. See Figure S4 for corresponding images of cells treated with hydrogen peroxide and paraquat. Scale bar, 2 μM. |
|
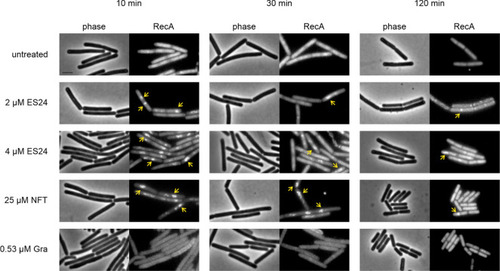
Localization of RecA-GFP (B. subtilis UG10) after treatment with ES24 or NFT. Gramicidin (Gra) was used as additional control. Yellow arrows indicate clustered protein. Scale bar, 2 μM. |
|
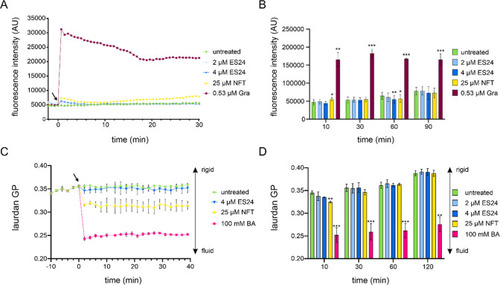
Effects of ES24 and NFT on B. subtilis membrane potential and fluidity. (A) Kinetic DiSC(3)5 spectroscopy measurements of B. subtilis 168CA treated with ES24, NFT, or positive control gramicidin (Gra). An increase in fluorescence intensity indicates membrane depolarization. The arrow indicates the time point of antibiotic addition. (B) End point measurements under the same conditions as in panel A up to 90 min. (C) Kinetic laurdan generalized polarization (GP) spectroscopy measurements of B. subtilis 168CA treated with ES24, NFT, or positive control benzyl alcohol (BA). A decrease in GP indicates membrane fluidization. The arrow indicates the time point of antibiotic addition. (D) End point measurements under the same conditions as in panel C up to 120 min. The p-values were calculated using a paired two-tailed t test. *p < 0.05, **p < 0.01, ***p < 0.001. |
|
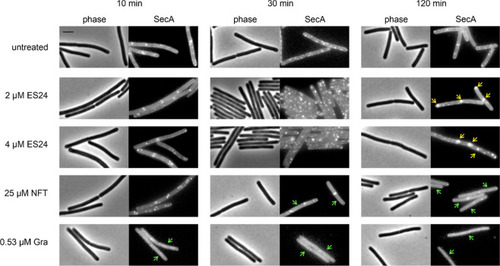
Localization of SecA-GFP after treatment with ES24 or NFT. Gramicidin (Gra) was used as additional control. Yellow arrows: clustered protein. Green arrows: dispersed protein. Scale bar, 2 μM. |
|
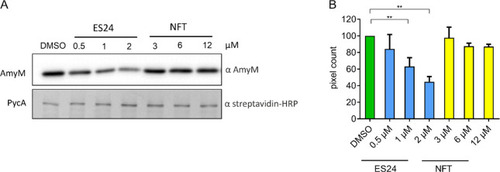
Effect on AmyM secretion. B. subtilis BWB09 (ΔxynA ΔamyE) carrying pCS73 (PamyQ–amyM) to express amyM from the constitutive PamyQ promoter was grown until mid-log phase and treated with different concentrations of ES24 or NFT for 3 h prior to SDS-PAGE and Western blotting. (A) Western blot analysis of culture supernatant using an α-AmyM antibody (top) and of cell lysate using an α-streptavidin antibody detecting the cytosolic protein PycA (bottom). (B) Quantification of pixel intensity in AmyM bands. **p < 0.01. |
|
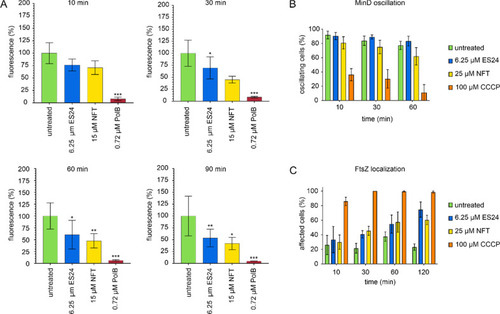
Effects of ES24 on cell membrane function in E. coli. (A) DiSC(3)5 measurements of E. coli MC4100 carrying pABCON2-fhuA ΔC/Δ4L treated with ES24 or NFT. Polymyxin B (PolB) was used as positive control. p-Values were calculated using a heteroscedastic, two-tailed t test. *p < 0.05, **p < 0.01, ***p < 0.001. (B) MinD oscillation in E. coli RC1 carrying pFX9 treated with ES24 or NFT. The proton ionophore CCCP, which is known to abolish MinD oscillation due to membrane depolarization,40 was used as positive control. (C) Quantification of FtsZ delocalization. Cells displaying an entirely cytosolic GFP signal, GFP clusters in the cell membrane, double bands, or spirals of FtsZ were counted as delocalized. Cells displaying single FtsZ bands, two opposing foci, or one focus at midcell (indicative of a closing septum) were counted as normal. |
|
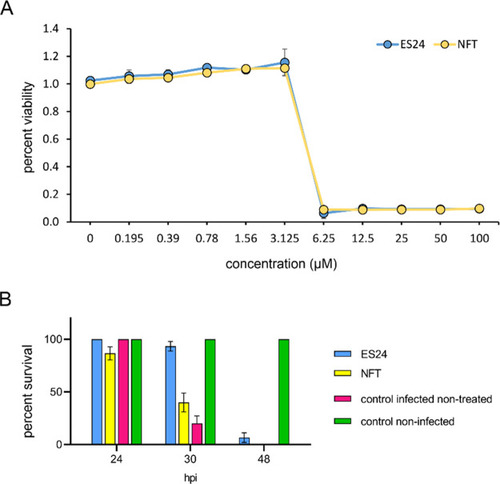
In vivo efficacy of ES24 in an S. pneumoniae zebrafish embryo infection model. (A) Viability of S. pneumoniae D39 after overnight incubation with a concentrations series of ES24 or NFT measured with resazurin. (B) Survival of 2 days postfertilization zebrafish embryos infected with S. pneumoniae D39. Embryos were injected with 150 CFU of bacteria in the caudal vein and subsequently treated with 5 μM ES24 or 5 μM NFT by adding the compounds directly to the water 1 h post infection (hpi). ES24 treatment improved the survival rate significantly in zebrafish embryos infected with S. pneumoniae D39 compared with NFT-treated embryos (p < 0.0001) or nontreated infected embryos (p < 0.0001). NFT also improved the survival rate compared with nontreated infected embryos, albeit not significantly (p = 0.4139). Data represents the mean ± SEM. Experiments were performed in biological triplicates with 10 fish per condition in each replicate. Survival rates were compared using log-rank statistics. |
|
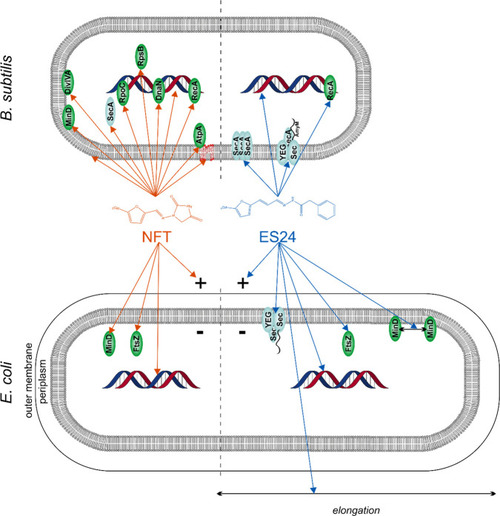
Current model of ES24 mechanism of action in Gram-positive and Gram-negative bacteria in comparison with NFT. NFT affects different cellular processes, including DNA packing; localization of DNA-binding proteins; and membrane integrity, membrane fluidity, and membrane protein binding. No clear differences were observed in its mechanism against E. coli and B. subtilis. ES24 affects Sec-mediated protein secretion and causes DNA damage in both organisms, but only affects cell division in E. coli. Reporter protein functions: MinD and DivIVA, cell division site regulation; FtsZ, cell division; SecA, Sec-dependent protein secretion; AtpA, ATP synthesis; RecA, DNA damage repair; DnaN, replication; RpoC, transcription; RpsB: translation. |