|
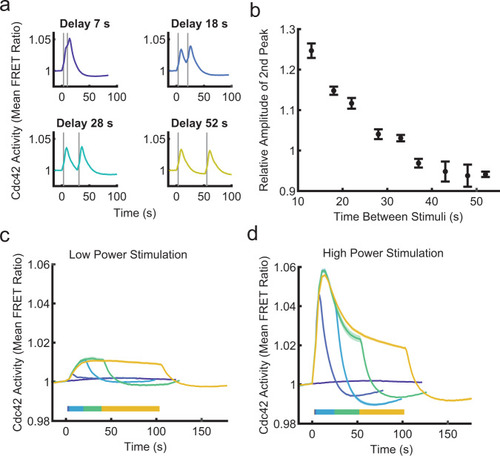
The Cdc42 circuit is regulated by complex, multi-level negative regulation.a Cdc42 responses are shown for four different two-pulse stimulation protocols with the indicated delay times between stimulations. The relative light intensity was ten for all plots. Data are presented as mean (nwell replicates = 30 for 7 s, nwell replicates = 6 for 18 s, nwell replicates = 6 for 28 s, and nwell replicates = 4 for 52 s). See Supplementary Fig. 2 for all two-pulse stimulation plots. Stimulus duration indicated by gray bars. b Relative amplitude of the second Cdc42 response peak as a function of time between stimuli. The relative amplitude is calculated as the ratio of the second peak to the first peak. A time delay of 7 s was not included because a second peak could not be resolved from the first. Data are presented as mean ± s.e.m. (nwell replicates = 6 for 13 s, nwell replicates = 6 for 18 s, nwell replicates = 6 for 22 s, nwell replicates = 6 for 28 s, nwell replicates = 26 for 33 s, nwell replicates = 4 for 37 s, nwell replicates = 4 for 43 s, nwell replicates = 4 for 48 s, and nwell replicates = 4 for 52 s). c, d Repeated global stimulations were applied to simulate continuous receptor stimulation. Stimulation duration is indicated by horizontal color bars. Data are presented as mean ± s.e.m. c Cdc42 response to the indicated stimulus durations with low-power stimulation (relative light intensity = 1). nwell replicates = 65 for non-stimulated, nwell replicates = 21 for 1 stimulation, nwell replicates = 20 for 7 stimulations, nwell replicates = 21 for 15 stimulations, and nwell replicates = 27 for 30 stimulations. d Cdc42 response to the indicated stimulus durations with high-power stimulation (relative light intensity = 100). nwell replicates = 65 for 0 stimulation condition, nwell replicates = 12 for 1 stimulation, nwell replicates = 12 for 7 stimulations, nwell replicates = 12 for 15 stimulations, and nwell replicates = 77 for 30 stimulations. Source data are provided as a Source Data file.
|