- Title
-
Insights into in vivo adipocyte differentiation through cell-specific labeling in zebrafish
- Authors
- Lepanto, P., Levin-Ferreyra, F., Koziol, U., Malacrida, L., Badano, J.L.
- Source
- Full text @ Biol. Open
|
|
|
|
|
|
|
|
|
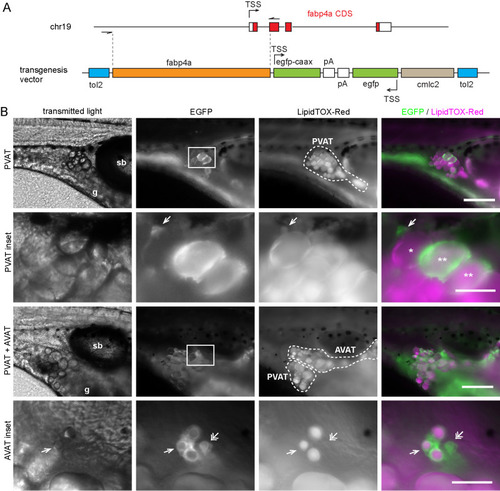
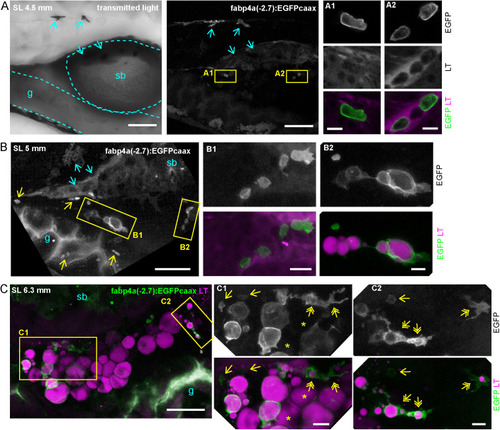
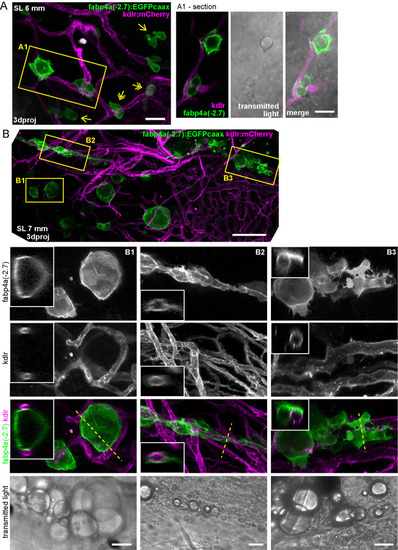
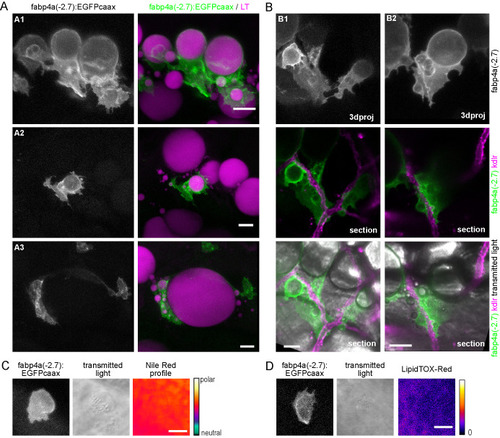
Different cell morphologies observed in fabp4a(-2.7):EGFPcaax larvae. (A) fabp4a(-2.7):EGFPcaax larvae of 21 dpf were labeled with LipidTOX-Red and imaged in vivo through confocal microscopy. Images are 3D projections of confocal stacks, to show different cell morphologies found in these larvae. (B) Images of fabp4a(-2.7):EGFPcaax; kdlr:mCherry larvae of SL 8 mm (19 dpf). Note labeled cells in the AVAT depot with cytoplasmic projections which lay in close apposition to blood vessels. Images are 3D projections or sections as indicated. (C,D) High magnification confocal sections of EGFP+ cells in larvae stained with Nile Red (C) or LipidTOX-Red (D). The cytoplasmic inclusions observed in transmitted light in these cells showed a polar lipid profile (evidenced by Nile Red staining, C) and low signal intensity of LipidTOX-Red (D). Scale bars: A: 20 μm; B: 20 μm; C,D: 10 μm. |
|
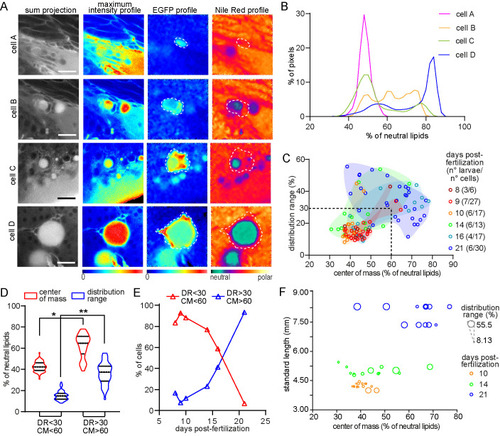
Larvae in different stages present cells with distinct lipid metabolic profiles. (A) Representative hyperspectral images of adipocytes (‘cell A’ to ‘cell D’) in different stages of differentiation. Raw images are presented in gray and intensity based color scale. Images generated after phasor plot analysis make evident the EGFP and Nile Red profiles which are represented separately by different color scales. Scale bars: A: 20 μm. (B) Plot with the normalized distribution of the image pixels with respect to the fraction (expressed as percentage) of neutral lipids for each cell in A. (C) Scatter plot showing the distribution range and center of mass of the cells analyzed in different larval stages. Both variables are expressed as the percentage of neutral lipids. The group of data coming from the same larval stage were enclosed by a manually generated convex hull shape just for visualization purposes. The dashed lines (DR=30; CM=60) generates the two regions considered in D and E. The numbers in brackets indicate the total number of larvae and the total number of cells analyzed in each stage. (D) Data within (DR<30/CM<60) and outside (DR>30/CM>60) the region considered in C were separated and compared. For CM: (*) P=2.06×10−13 for median comparison (Mann–Whitney test), P=1.62×10−4 for coefficient of variation comparison (Fligner-Killeen test); (**) P=2.24×10−16 for median comparison (Mann–Whitney test), P=7.1×10−6 (Fligner-Killeen test). (E) Representation of the percentage of cells in each group (DR<30/CM<60 and DR>30/CM>60) with respect to the larval stage presented in dpf. (F) Representation of the distribution range (as the size of the dots) and center of mass for the cells within some of the larva analyzed; each larva had a different standard length. |