- Title
-
In vivo validation of a computationally predicted conserved Ath5 target gene set
- Authors
- Del Bene, F., Ettwiller, L., Skowronska-Krawczyk, D., Baier, H., Matter, J.M., Birney, E., Wittbrodt, J.
- Source
- Full text @ PLoS Genet.
|
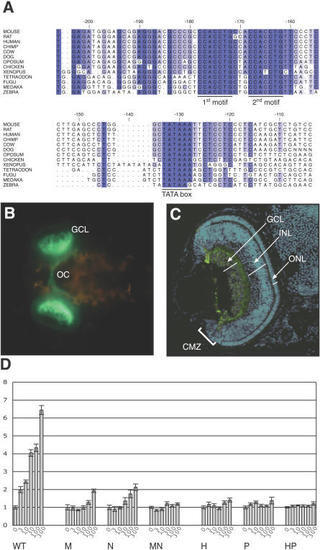
(A) Proximal promoter region alignment of (B, C) Transgenic medaka embryos at day four of development. (C) Cryosection and anti-GFP antibody staining of a transgenenic embryo representing the full Ath5 expression pattern. Note that the reporter is weakly expressed all over the mature retina and specifically enriched in the GCL and in a few other cells indicated by arrows. Complete absence of expression is observed in the distal part of the ciliary marginal zone as well as outside the eye. Nuclei are visualized by DAPI counterstaining. CMZ, ciliary marginal zone; GCL, ganglion cell layer; INL, inner nuclear layer; ONL outer nuclear layer; and OC, optic chiasm. (D) Transactivation of Ath5 on its own promoter assayed by luciferase reporter transcription activity. Two-kilobase Ath5 promoter or its mutant forms generated by PCR introducing base changes into the two conserved binding motifs were used (see |
|
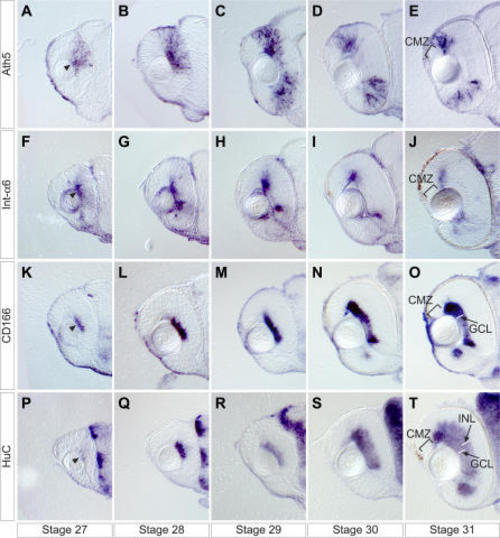
Embryos are stained by whole mount in situ hybridization and sectioned transversally at the level of the optic nerve. |
|
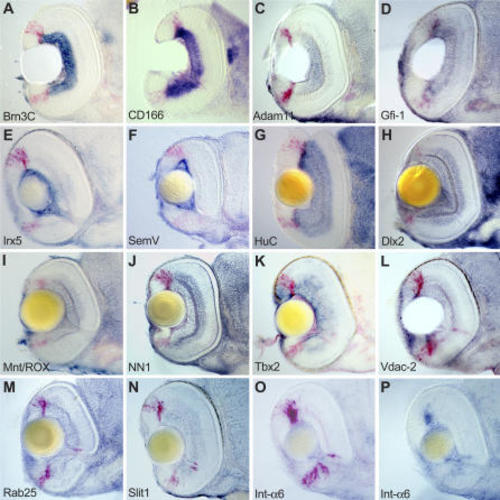
(A–P) Embryos are sectioned transversally at the level of the optic nerve. Dorsal is to the top. Abbreviations of target genes are indicated in |
|
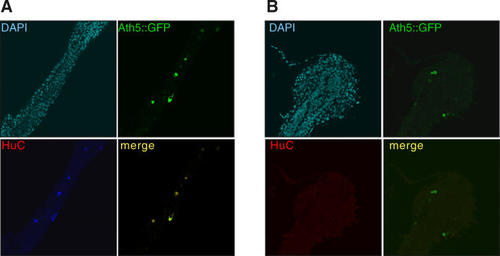
(A, B) Mosaic expression of Ath5 leads to activation of target genes in those cells expressing Ath5. Upper left, DAPI nuclear staining; lower left, HuC fluorescent in situ; upper right, GFP reporter indicating Ath5 expression; and lower right, merge of GFP and in situ signals. (A) Medaka embryo injected with 2-kb Ath5 promoter driving GFP expression in response to Ath5 and Ath5 pCS2+ expression vector. Note the complete overlap of Ath5 activity and expression of its target HuC. (B) Medaka embryo control injected with 2-kb Ath5 promoter driving GFP expression and empty pCS2+ vector. |
|
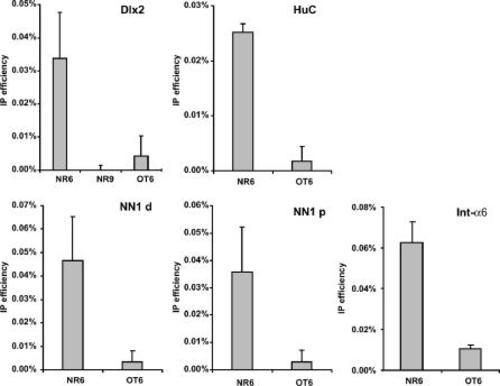
Antibody directed against Ath5 was used to immunoprecipitate crosslinked chromatin fragments prepared from chicken neuroretina and optic tectum. Occupation was measured by real time PCR with primers specific for the predicted target genes ( |