- Title
-
REST is a major negative regulator of endocrine differentiation during pancreas organogenesis
- Authors
- Rovira, M., Atla, G., Maestro, M.A., Grau, V., García-Hurtado, J., Maqueda, M., Mosquera, J.L., Yamada, Y., Kerr-Conte, J., Pattou, F., Ferrer, J.
- Source
- Full text @ Genes & Dev.
|
|
|
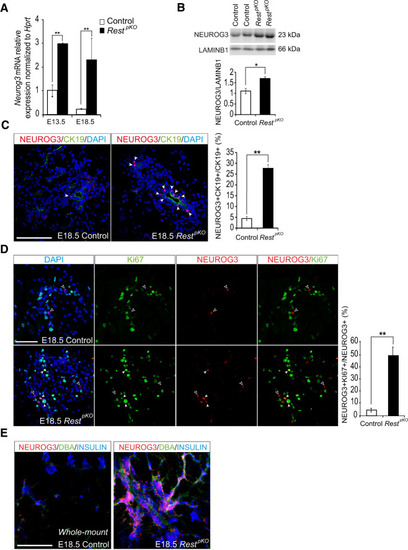
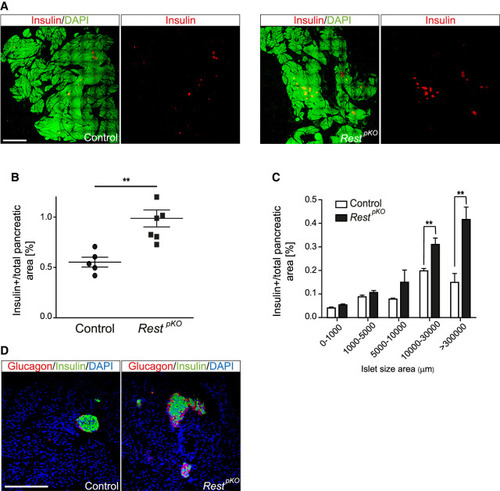
Increased β-cell mass in |
|
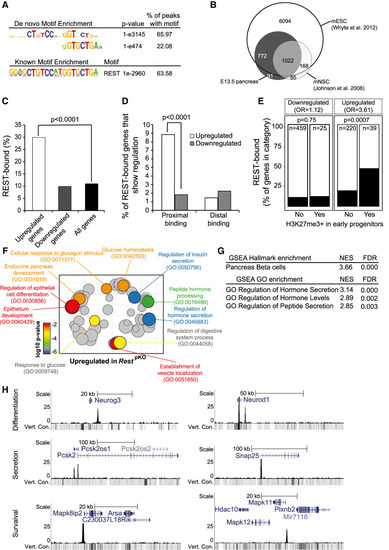
Functional direct REST targets in the embryonic pancreas. (A) Top de novo and known motif enrichments in REST-bound regions. (B) REST-bound regions in E13.5 pancreas, mESCs, and mNSCs. (C) Percentage of up-regulated and down-regulated genes in RestpKO mice, or all genes, that were bound by REST. Results indicate that REST predominantly acts as a repressor. P-values are Fisher exact test. (D) REST binds preferentially to promoter-proximal (0- to 5-kb) regions of genes that were up-regulated in RestpKO mice. (E) Percentage of differentially expressed genes that were bound by REST, broken down by H3K27me3 enrichment in purified pancreatic progenitors in (van Arensbergen et al. 2010). REST binding was enriched in genes that were up-regulated in RestpKO and showed H3K27me3 in progenitors. P-values from Fisher exact test. (F) Up-regulated genes were functionally annotated using Gorilla (Eden et al. 2009), and REVIGO (Supek et al. 2011) was used to visualize annotation clusters. The most significant terms are highlighted according to a P-value color scale. (G) Significant GSEA terms for up-regulated genes. (H) REST binding associated to pancreatic endocrine development (Neurog3 and NeuroD1), insulin secretion (Snap25 and Pcsk2), and β-cell survival (Mapk11 and Mapk8ip2) genes in E13.5 pancreas. Y-axes are −log10 P-values. The vert. cons. track depicts vertebrate conservation. |
|
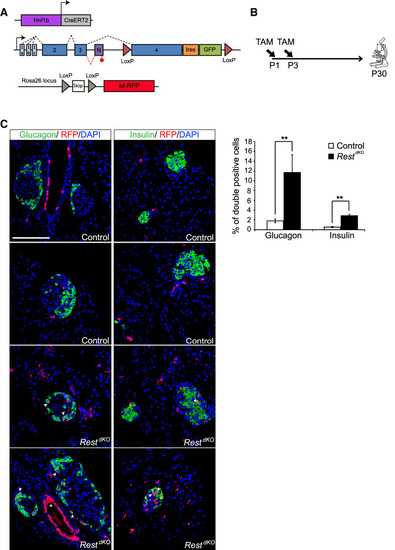
Pancreas-specific inactivation of Rest in neonatal ducts. (A) Schematic of genetic models used to inactivate Rest and activate RFP expression in duct cells and progeny. Hnf1b-CreERT2 is a BAC transgenic that specifically marks duct cells (Solar et al. 2009), as well as non-Rest-expressing ∂ cells in reporters that are excised with high efficiency (Rovira et al. 2021), but not other endocrine or acinar cells. (B) Schematic of the lineage tracing experiment. Tamoxifen was given to mothers at day 1 (P1) and day 3 (P3) after delivery, and mice were analyzed at P30. Hnf1b-CreERT2;Rosa26RFP control mice were also treated. (C) Representative images of double-positive RFP (red) and insulin (green) cells and of double-positive RFP (red) and glucagon (green) cells in RestdKO and control mice. The graph shows RFP-expressing glucagon and insulin cells in RestdKO versus control mice. n = 5–6 mice per each group. Arrowheads indicate double-positive cells, and an asterisk marks examples of cells in a duct, which were very efficiently labelled. Scale bar, 100 µm. Error bars are SEM. Student's t-test; (**) P < 0.01. |
|
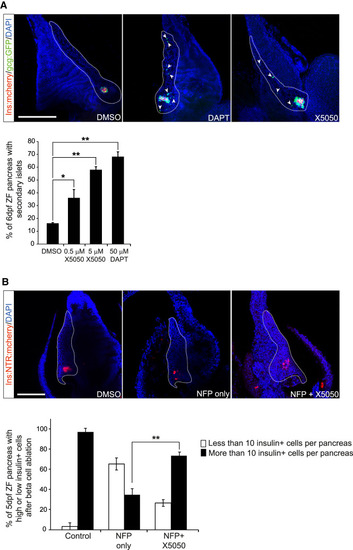
REST regulation of endocrine differentiation is conserved in zebrafish. ( |
|
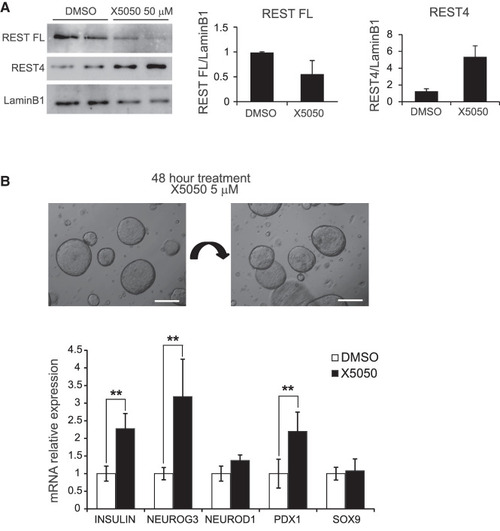
REST chemical inhibition in human pancreatic organoids. ( |