- Title
-
Origin and evolution of the zinc finger antiviral protein
- Authors
- Gonçalves-Carneiro, D., Takata, M.A., Ong, H., Shilton, A., Bieniasz, P.D.
- Source
- Full text @ PLoS Pathog.
|
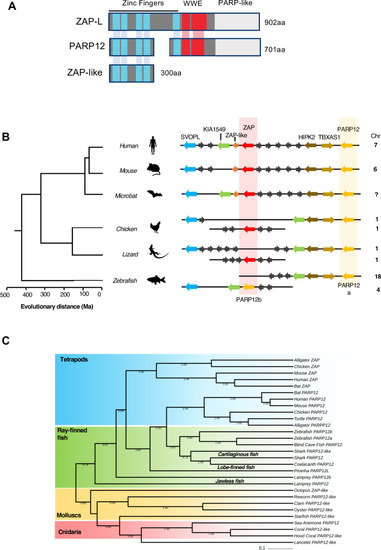
(A) Schematic representation of the domain organization of human ZAP and its paralogues PARP12 and ZAP-like. (B) Diagrams of the organization of PARP12 (in yellow) and ZAP (in red) loci in vertebrate genomes generated using multiple genome browsers (NCBI, Ensembl, Genomicus). Evolutionary distance of the indicated species (presented in million years, Ma) is based on [ |
|
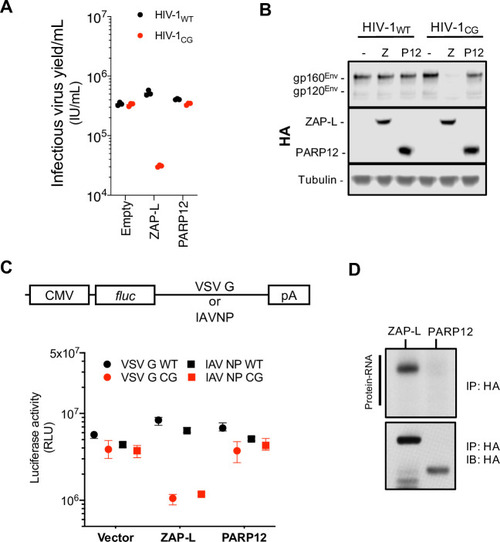
(A-B) HEK293T ZAP-/- cells were transfected with HIV-1WT or HIV-1CG plasmids and plasmids encoding human ZAP-L-HA or PARP12-HA. Supernatant was harvested after 48h and the infectious virus yield was determined using MT4-R5-GFP target cells (A). Whole cell lysates were analysed by western blotting (B). IU, infectious units. (C) HEK293T ZAP-/- cells were transfected with plasmids encoding a luciferase reporter gene that contained VSV-G wildtype (WT) or CG-enriched (VSV-G CG) sequences, or IAV-NP WT or CG-enriched sequences as 3’ UTRs, together with plasmids expressing ZAP-L, PARP12 or an empty plasmid (vector). RLU, relative light units. (D) HEK293T ZAP-/- cells were transfected with plasmids expressing ZAP-L-3xHA or PARP12-3xHA and 24h later culture medium was supplemented with 100μM 4SU. After overnight incubation cells were irradiated with UV light, and ZAP-L/PARP12 were immunoprecipitated. RNA bound to each protein was radiolabeled and protein-RNA adducts were resolved by SDS-PAGE, transferred to a nitrocellulose membrane and exposed to autoradiographic film. In parallel, a western blot of immunoprecipitated proteins was done using anti-HA antibody. IP, immunoprecipitation. IB, immunoblot. |
|
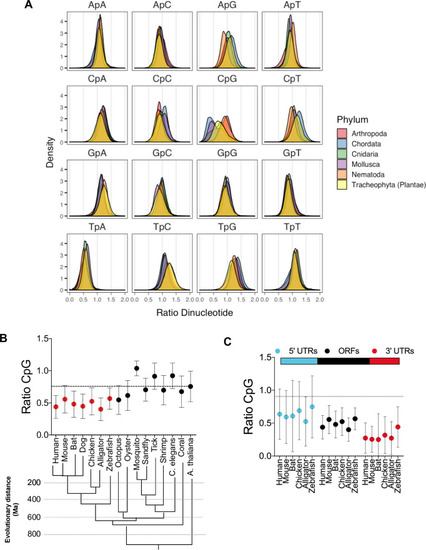
(A) Open reading frames (ORFs) from several organisms belonging to the indicated phyla were collected from the NCBI nucleotide database and dinucleotide frequency ratio (observed/expected) was calculated and frequency distribution for each dinucleotide in ORFs was plotted. (B) Average dinucleotide observed/expected ratio in various animal species. Evolutionary distance (presented in million years, Ma) of indicated species was based on [ |
|
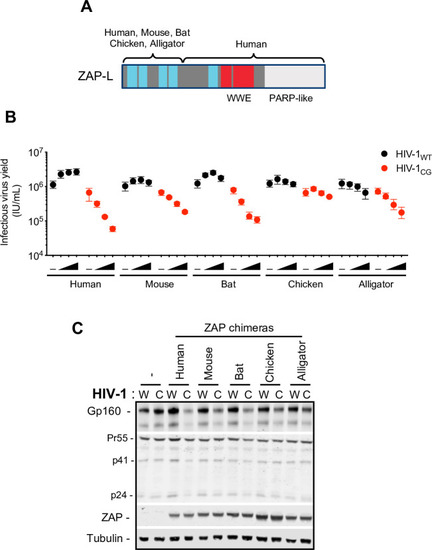
(A) Schematic representation of ZAP chimeras in which the RNA binding domain of mouse, bat, chicken and alligator ZAP was fused to the WWE and PARP-like domain of human ZAP. (B-C) HEK293T ZAP-/- cells were transfected with HIV-1WT (W) or HIV-1CG (C) proviral plasmids and increasing amounts (0ng, 75ng, 145ng, or 290ng) of a plasmid encoding each ZAP chimera. Supernatant was harvested after 48h and the infectious virus yield was determined using MT4-R5-GFP target cells (B). Whole cell lysates corresponding to the highest amount of ZAP plasmids transfected were analysed by western blotting probing with antibodies against HIV-1 proteins and ZAP (C). -, Indicates co-transfection with an empty vector in place of a ZAP expression plasmid. |
|
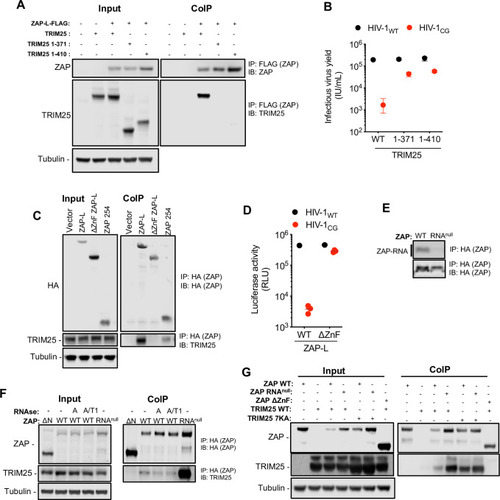
(A) HEK293T ZAP-/- and TRIM25 -/- cells were co-transfected with plasmids expressing human ZAP-L-FLAG and full-length untagged human TRIM25 or one of two human TRIM25 truncations (corresponding to the N-terminal 371 or 410 amino acids of TRIM25), that lack the SPRY domain [ |
|
(A) Phylogenetic analysis of TRIM25 protein sequences from vertebrate species. Shaded areas indicate cluster sequences from mammals (in blue), birds and reptiles (in green) and ray-finned fish (in yellow). (B) HEK293T ZAP-/- and TRIM25-/- cells were co-transfected with an HIV-1CG proviral plasmid as well as a fixed amount of a plasmid encoding human ZAP or ZAP chimeras from various vertebrate species. Animal names indicate source of the RNA-binding domains for each ZAP chimera. For each ZAP protein, cells were also co-transfected increasing amounts of a plasmid (0ng 10ng, 30ng or 90ng) expressing a TRIM25 protein from the 6 different indicated species. After 48h, infectious virus yield was determined using MT4-R5-GFP target cells. |
|
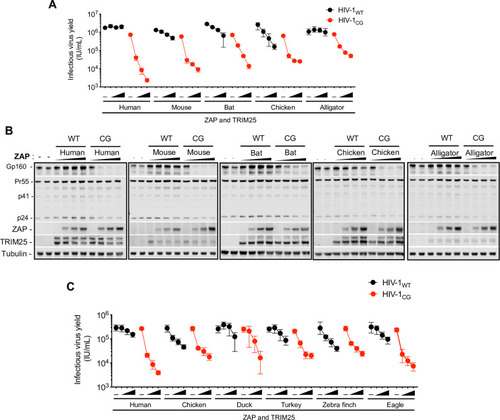
(A-B) HEK293T ZAP-/- and TRIM25-/- cells were co-transfected with indicated proviral plasmids, increasing amounts (0ng, 75ng, 145ng, or 290ng) of a plasmid encoding an FLAG-tagged human ZAP or mouse, bat, chicken and alligator ZAP chimeras and a fixed amount of a plasmid encoding a cognate HA-TRIM25 protein. After 48h, infectious virus yield was determined using MT4-R5-GFP target cells (A). Whole cell lysates were analysed by western blotting probing with antibodies against HIV-1 proteins ZAP-FLAG and TRIM25-HA (B). (C) HEK293T ZAP-/- and TRIM25-/- cells were transfected with HIV-1 proviral plasmids, along with increasing amounts (0ng, 75ng, 145ng, or 290ng) of a plasmids expressing human ZAP or ZAP chimeras from various avian species. For cells transfected with human ZAP, a fixed amount of a human TRIM25 expression plasmid was co-transfected, while for the avian ZAP chimeras, chicken TRIM25 was used. After 48h, infectious virus yield was determined using MT4-R5-GFP target cells. |
|
(A) HEK293T ZAP-/- and TRIM25-/- cells were transfected with an HIV-1CG proviral plasmid and a plasmid expressing human ZAP or chicken ZAP chimera. Cells were treated with 4-thiouiridine prior to UV-crosslinking and CLIP analysis. Reads associated with human (top) or chicken (bottom) ZAP were mapped to the HIV-1CG genome and the read density plotted against position in the HIV-1CG genome. CpG-enriched region the HIV-1CG genome is highlighted in red and dashed lines. (B) Contribution (% observed/expected) for each dinucleotide to reads derived from the HIV-1CG genome to was calculated. Shift in the ratio for each dinucleotide contribution comparing human and chicken ZAP reads was also determined (blue line). |
|
(A) Representation of the crystal structure of the RNA-binding NTD domain of human ZAP (PDB 6UEI, [ |