- Title
-
Cell Proliferation and Collective Cell Migration During Zebrafish Lateral Line System Development Are Regulated by Ncam/Fgf-Receptor Interactions
- Authors
- Dries, R., Lange, A., Heiny, S., Berghaus, K.I., Bastmeyer, M., Bentrop, J.
- Source
- Full text @ Front Cell Dev Biol
|
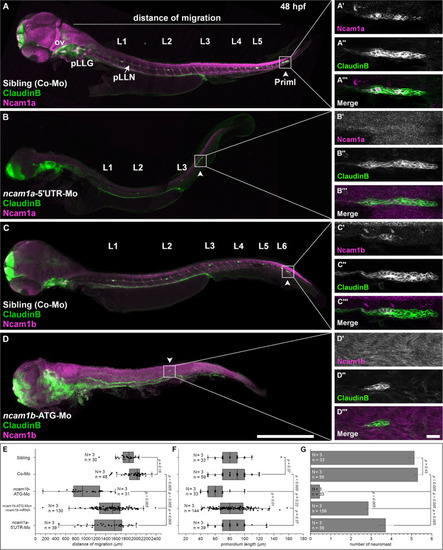
Expression pattern and function of Ncam1 during development of the zebrafish lateral line system. |
|
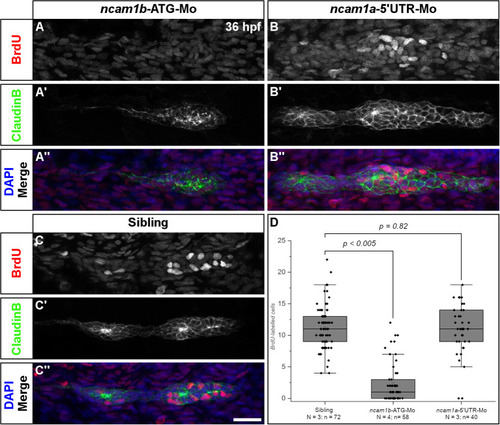
Ncam1b is required for cell proliferation within the primordium. Siblings or morpholino-injected |
|
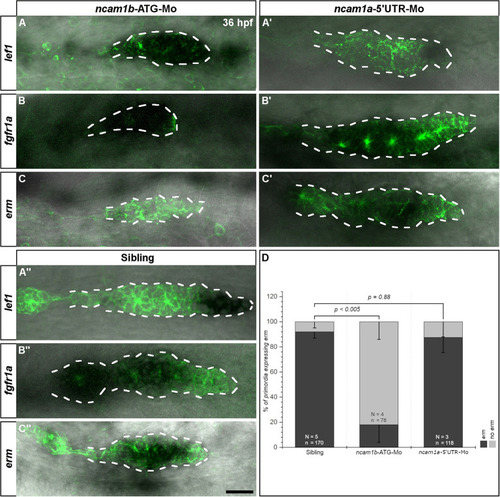
Ncam1b affects the expression of the Fgfr1a-target gene |
|
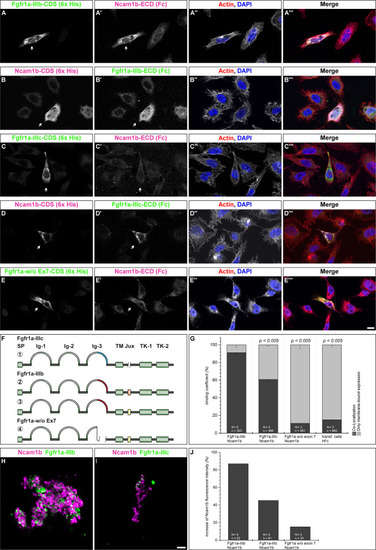
Ncam1b interactions with different isoforms of Fgfr1a. |
|
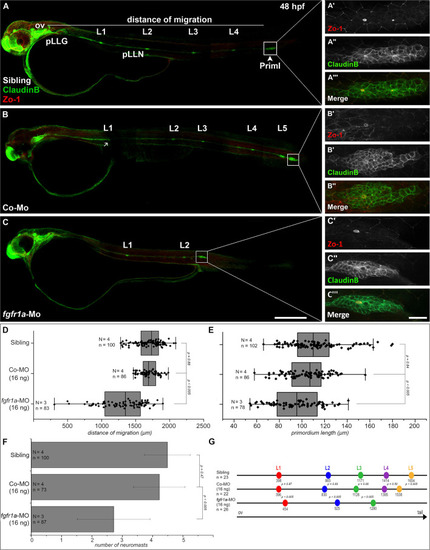
Fgfr1a is important for posterior lateral line development. Lateral views of 48 hpf |
|
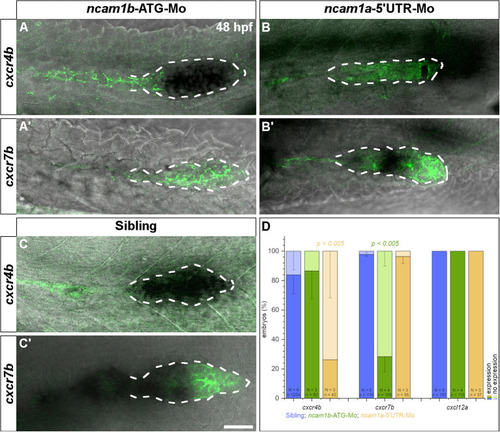
Expression of chemokine receptor |
|
The function of Ncam1b during posterior lateral line development. |