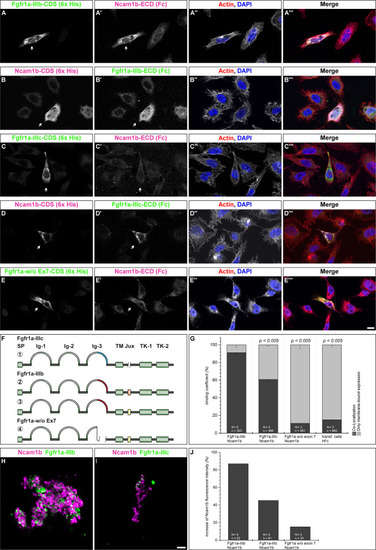
Ncam1b interactions with different isoforms of Fgfr1a. CHO-K1 cells were transfected with complete coding sequences (CDS) of one of three Fgfr1a-isoforms (green; IIIb, IIIIc, or w/o Ex7) or of Ncam1b (magenta). Transfected cells were incubated for 1 hr with either soluble Ncam1b or one of the two Fgfr1a-isoforms, fixed and immunostained for His-, Fc-Tag, Actin (red), and DAPI (blue). (A,B‴) Ncam1b interacts strongly with Fgfr1a-IIIb irrespectively of which of the proteins is membrane-bound. (C,D‴) Binding of Ncam1b to Fgfr1a-IIIc is markedly weaker. (E,E‴) No interaction occurs if Fgfr1a lacks exon 7. (F) Scheme of isolated and cloned Fgfr1a splice variants: ① Fgfr1a-IIIc does not contain any exon 9, ② one splice variant of Fgfr1a-IIIb contains a juxtamembrane region of valine and threonine (orange), ③ another isoform of Fgfr1a-IIIb encodes two serine residues (yellow) within the juxtamembrane domain, ④ Fgfr1a-IIIa, an artificial variant of Fgfr1a, lacks the C-terminal part of Ig-3, which is encoded by exon 7. (G) Quantification of the binding of Ncam1b to splice variants of Fgfr1a. Binding coefficient represents the percentage of cells expressing the membrane-bound binding partner that were co-labeled by antibodies against the soluble binding partner. Error bars represent standard deviations (χ2-test). (H,I) Bead aggregation assays reveal a strong homophilic binding of Ncam1b. Ncam1b-coated beads interact with Fgfr1a-IIIb-coated beads and to a lower extend with Fgfr1a-IIIc. (J) Measuring the fluorescence intensity of soluble Ncam1b-Fc shows a strong binding to membrane-bound Fgfr1a-IIIb and a weaker to Fgfr1a-IIIc or Fgfr1a without exon 7. Scale bar in (E‴) represents 20 μm and in (I) 10 μm. Abbreviations: CDS, coding sequence; ECD, extracellular domain; lg, Immunoglobulin domain; Jux, juxtamembrane domain; SP, Signal peptide; TK, Tyrosine Kinase domain.
|

