- Title
-
Osmolarity-independent electrical cues guide rapid response to injury in zebrafish epidermis
- Authors
- Kennard, A.S., Theriot, J.A.
- Source
- Full text @ Elife
|
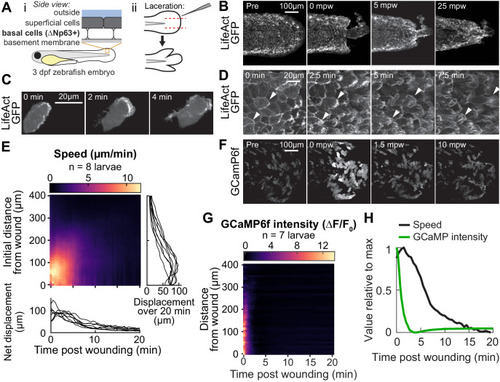
(A) Schematic of (i) bilayered larval zebrafish skin and (ii) laceration technique. (B) Lacerated tailfin over time from a larva 3 days post fertilization (dpf) expressing LifeAct-EGFP in basal cells (TgBAC(∆Np63:Gal4); Tg(UAS:LifeAct-EGFP); Tg(hsp70:myl9-mApple)). mpw: minutes post wounding. (B–F) are all maximum-intensity Z-projections of spinning-disk confocal images. (C) Individual cell from 3 dpf larva expressing LifeAct-EGFP mosaically in basal cells (TgBAC(∆Np63:Gal4) larva injected with UAS:LifeAct-EGFP plasmid at the 1-cell stage). Wound was to the right approximately 1–2 min earlier. (D) Cells in a lacerated tailfin over time from 3 dpf larva expressing LifeAct-EGFP in basal cells (TgBAC(∆Np63:Gal4); Tg(UAS:LifeAct-EGFP); Tg(hsp70:myl9-mApple)), approximately 1–2 min post wounding. Arrowheads: examples of individual actin-rich protrusions are followed over time. (E) Kymograph indicating the speed of basal cells at a given distance from the wound over time (N = 8 larvae). Line graphs show net displacement over space (right) and time (bottom) for each individual larva. See Methods and Figure 1—figure supplement 1 for details of motion tracking analysis. (F) Lacerated tailfin from larva expressing GCaMP6f in basal cells (TgBAC(∆Np63:Gal4) larvae injected with UAS:GCaMP6f-P2A-nls-dTomato plasmid at the 1 cell stage). mpw: minutes post wounding. Due to the large dynamic range in GCaMP intensity, these images were gamma-corrected with a gamma of 0.5 for display purposes. (G) Kymograph of GCaMP6f intensity, normalized by the coexpressed nuclearly localized dTomato intensity, and relative to the normalized intensity pre-wounding (F0) (N = 7 larvae). (H) Line graph of normalized profiles of the average speed and GCaMP intensity over time, averaged over 300 µm of tissue closest to the wound. To emphasize comparison of the temporal relationship, profiles are rescaled to lie between 0 and 1 (in arbitrary units). |
|
( |
|
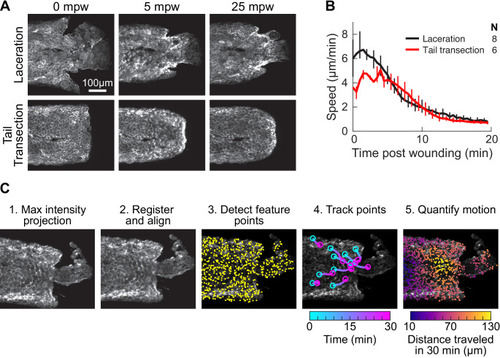
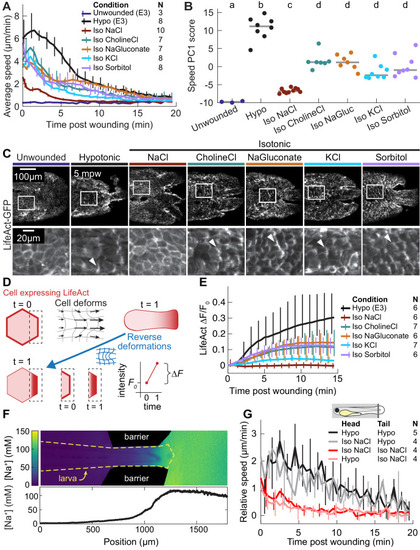
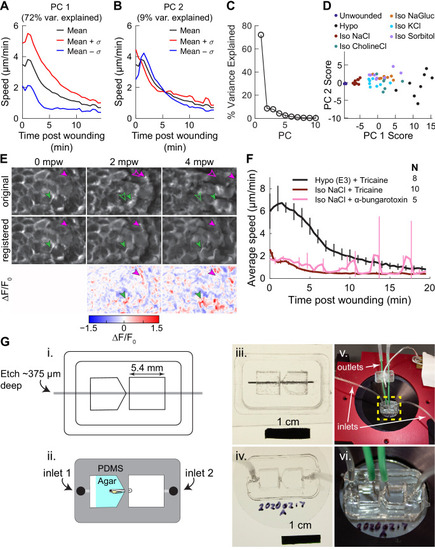
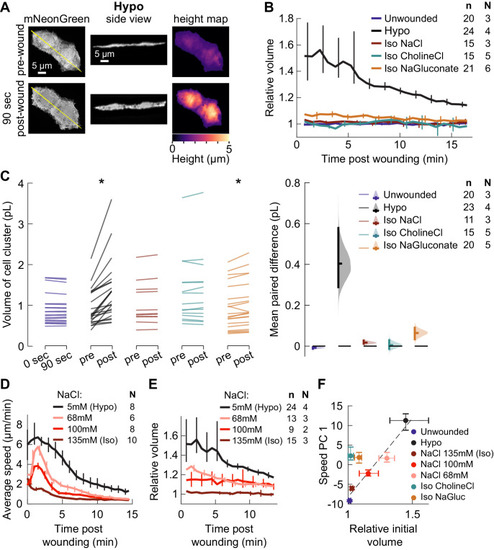
(A) Basal cell speed over time, averaged over 300 µm adjacent to the wound in each larva. 3 dpf larvae expressing LifeAct-EGFP in basal cells (TgBAC(∆Np63:Gal4); Tg(UAS:LifeAct-EGFP); Tg(hsp70:myl9-mApple)) were incubated in E3 (Hypo) or E3 supplemented with 270 mOsmol/l of indicated osmolytes (Iso) and then the tailfin was lacerated and movement analyzed as described in Methods and Figure 1—figure supplement 1. N indicates the number of larvae in each condition. Error bars are bootstrapped 95% confidence intervals of the mean for each condition. (B) Speed trajectories for each larva were analyzed with PCA (see Figure 2—figure supplement 1A–C) and each trajectory’s score along the first principal component is plotted. Gray bars indicate the mean PC1 score for that condition. Letters a-d indicate statistically distinguishable (significantly different) means (p<0.001, one-way fixed-effects Welch’s ANOVA F(6, 19)=130.9, with Games-Howell post-hoc tests). See Table 1 for p-values from post-hoc tests. (C) (Top) Representative tailfins from unwounded larvae or larvae wounded in different media. Images shown from 5 min post wounding. (Bottom) Insets shown below each image. Arrowheads: examples of polarized LifeAct intensity, in the direction of the wound. (D) Schematic of computational procedure for analyzing changes in intensity, after warping image to account for cell/tissue deformation. See Methods for more detail. (E) Relative pixel-wise change in LifeAct intensity over time, averaged over 300 µm adjacent to the wound in each larva. Error bars are bootstrapped 95% confidence intervals of the mean. (F) (Top) Image displaying the device allowing for different media compositions around the tailfin or the rest of the larva. Sodium concentration was calibrated with a sodium-sensitive fluorescent dye. (Bottom) Graph indicates the average sodium concentration along a line across the middle of the image. (G) Relative tissue speed for larvae with different media around their anterior or posterior, as shown in the diagram. To account for residual whole-larva movement due to peristaltic flow, the average tissue speed >300 µm away from the wound was subtracted from the average speed <300 µm away from the wound. Error bars are bootstrapped 95% confidence intervals of the mean. |
|
( |
|
( |
|
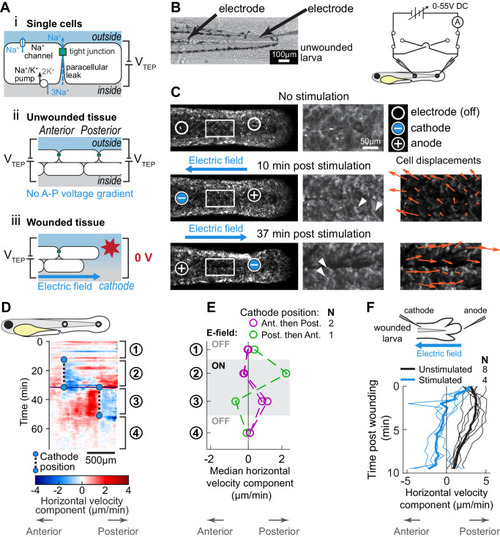
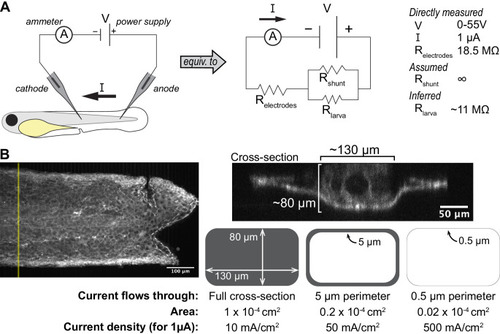
(A) (i) Schematic of the origin of the transepithelial potential (TEP) due to circulating flow of sodium ions. Other ions (such as chloride) may also be transported using energy derived from the sodium-potassium pump, and are not shown here for clarity. (ii) Unwounded tissue does not show an anterior-posterior TEP gradient. (iii) Wounding short-circuits TEP leading to an anterior-posterior TEP gradient and electric field. (B) (Left) Brightfield image of tailfin from 3 dpf larva expressing LifeAct-EGFP in basal cells (TgBAC(∆Np63:Gal4); Tg(UAS:LifeAct-EGFP)), with electrodes inserted under the skin. (Right) Electrical stimulation circuit with variable DC voltage, current measurement, and switches to reverse current polarity in the larva. (C) Z-projections of LifeAct signal from larva shown in (B). (Top) electric field off; (Middle) electric field on with cathode at anterior electrode; (Bottom) electric field reversed with cathode at posterior electrode. Stills are from one continuous timelapse. Insets are shown, and displacement vectors from tissue motion tracking are shown in orange. Arrowheads: examples of polarized LifeAct intensity oriented toward the cathode. (D) Velocity kymograph from a representative timelapse. Color indicates horizontal velocity component from tissue motion tracking analysis. Blue circles and dashed lines indicate the position of the cathode when the electric field was turned on, roughly corresponding to the empty circles on the larva diagram. Numbers 1–4 indicate different phases of the timelapse. 1: electric field off; 2: electric field on, cathode anterior; 3: electric field on, cathode posterior; 4: electric field off. (E) Median horizontal velocity component from three different larva. Tissue velocity was averaged in the region between the two electrodes and then the median velocity during each phase 1–4 (described above) was plotted. For one larva (shown in green) the cathode was initially positioned at the posterior electrode and was then switched to the anterior electrode. (F) Average horizontal velocity component from stimulated or unstimulated larvae. In the stimulated condition, 3 dpf larvae expressing LifeAct-EGFP in basal cells (TgBAC(∆Np63:Gal4); Tg(UAS:LifeAct-EGFP); Tg(hsp70:myl9-mApple)) were impaled with one electrode, with the other electrode positioned immediately posterior to the tailfin. Larvae were then wounded and the electric field turned on, with the cathode positioned at the anterior electrode. Tissue motion between the cathode and the wound was analyzed. Thin lines represent velocity for each larva. Thick lines represent average over larvae. Unstimulated data is the same as in Figure 2A. |
|
( |