- Title
-
The DNA methyltransferase DNMT3A contributes to autophagy long-term memory
- Authors
- González-Rodríguez, P., Cheray, M., Füllgrabe, J., Salli, M., Engskog-Vlachos, P., Keane, L., Cunha, V., Lupa, A., Li, W., Ma, Q., Dreij, K., Rosenfeld, M.G., Joseph, B.
- Source
- Full text @ Autophagy
|
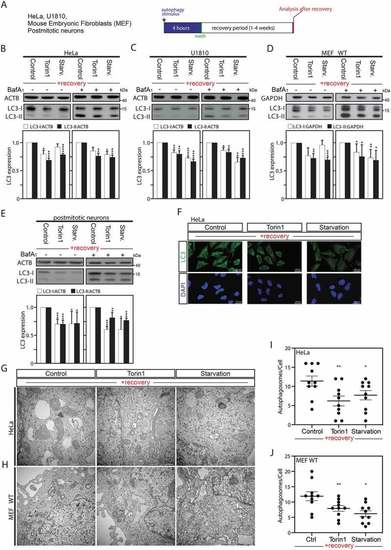
Autophagy induction is associated with a sustained downregulation of MAP1LC3 protein expression after a recovery period. (A) Illustrative pipeline of the “previously-autophagy exposed cells” generation and analysis after a recovery period. (B–E) Immunoblot analysis and quantification of LC3-I and LC3-II expression versus ACTB or GAPDH in HeLa cells (B), U1810 cells (C), wild-type (WT) MEF cells (D), or post-mitotic neurons (E), starved (Starv.), pretreated with torin1, or DMSO (used as control) for 4 h and thereafter left to recover and analyze after 2 weeks (B and C), one week (D), or 8 d (E) under normal cell culture conditions. Treatment with the late inhibitor of autophagy bafilomycin A1 (BafA1) before sample collection validates that the observed overall decrease in LC3 expression was not the result of an increase in autophagic flux (B to E). (F) Immunofluorescence confocal microscopy imaging of endogenous LC3 in HeLa cells after 2-week recovery period. (G to J) Ultrastructural analysis by electron microscopy of HeLa cells (G and I) or MEF cells (H and J), starved, pretreated with torin1, or DMSO (used as control) for 4 h and thereafter left to recover and analyze after 2 weeks (HeLa cells), or one week (MEF cells). Panels I and J are quantification of number of autophagosomes per cell. 10 cells were counted per conditions. All values are a mean of at least 3 independent experiments ± SEM and considered significant for *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. n.s., not significant for the indicated comparison. (B-E, n = 3–5; I-J, n = 10) |
|
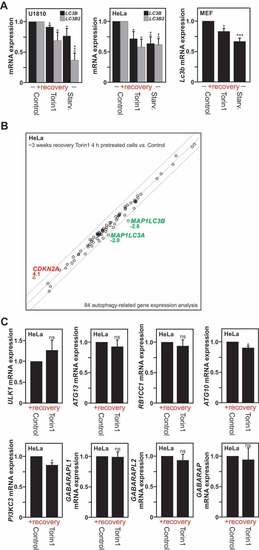
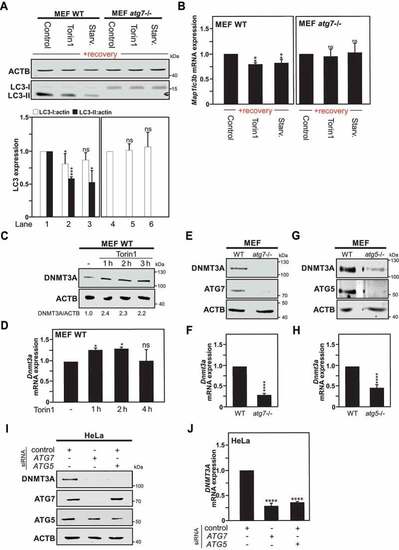
Transcriptional repression of the |
|
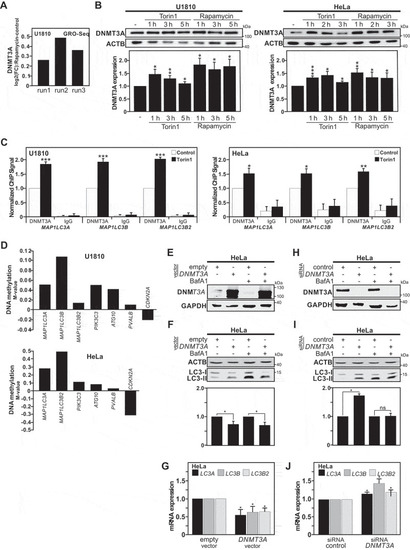
DNMT3A recruitment and DNA methylation occur at the |
|
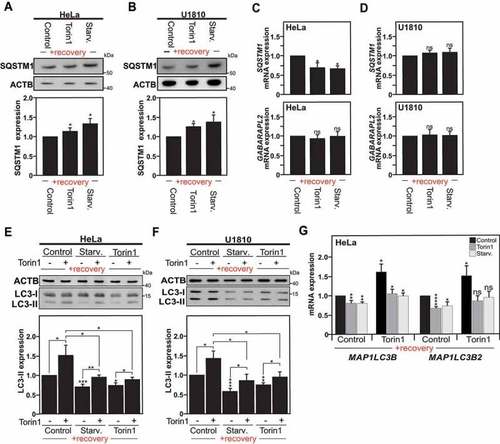
Autophagy is altered in cells previously exposed to an autophagy stimulus. (A and B) Immunoblot analysis of SQSTM1 expression in Hela cells (A), or U1810 cells (B), starved (Starv.), treated with torin1 or DMSO (used as control) for 4 h, and left to recover under normal culture conditions for a two to three weeks period. The graphs show the quantification of SQSTM1 versus ACTB expression in Hela cells (A), and U1810 cells (B). (C and D) SQSTM1 and GABARAPL2 mRNA expression levels measured by RT-qPCR in HeLa cells (C) and U1810 cells (D) previously exposed to an autophagy stimulus or DMSO (used as control) and analyzed at 2 weeks after the first autophagy stimulus. (E and F) Immunoblot and quantification analysis of LC3-II expression in Hela cells (E), or U1810 cells (F), previously exposed to an autophagy stimulus as described in Fig. S1A upon re-stimulation of autophagy with torin1 treatment for 1 h (+) as compared to DMSO treatment used as control (-). G MAP1LC3B and MAP1LC3B2 isoforms mRNA expression measured by RT-qPCR in HeLa cells previously exposed to an autophagy stimulus upon re-stimulation of autophagy with torin1. All values are means of at least 3 independent experiments ± SEM and considered significant for *p < 0.05, **p < 0.01, ***p < 0.001. n.s., not significant for the indicated comparison. (n = 3–6) |
|
MAP1LC3 downregulation is abrogated in |
|
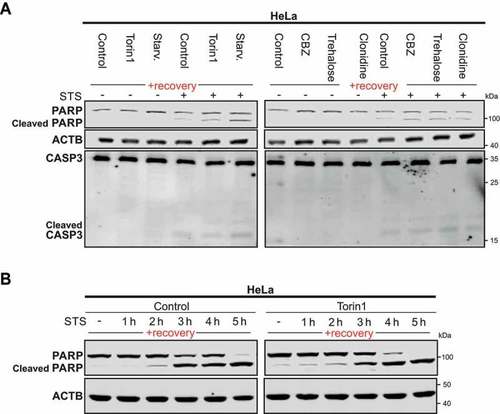
Previously-autophagy exposed cells are sensitized to apoptosis. (A) Representative immunoblot of 3 independent experiments of PARP and CASP3 in Hela cells starved (Starv.), treated with torin1 or MTOR-independent inducers for 4 h, thereafter left to recover for 2 weeks under normal cell culture conditions and treated with staurosporine (STS) for 3 h before harvest. CBZ., carbamazepine. (B) Representative immunoblot of 3 independent experiments of PARP and cleaved-PARP immunoblot of HeLa cells treated for 4 h with DMSO (used as control) and torin1 and left to recover for 2 weeks as previously described in Fig. S1A. Cleaved PARP was analyzed after STS treatment at different time points |
|
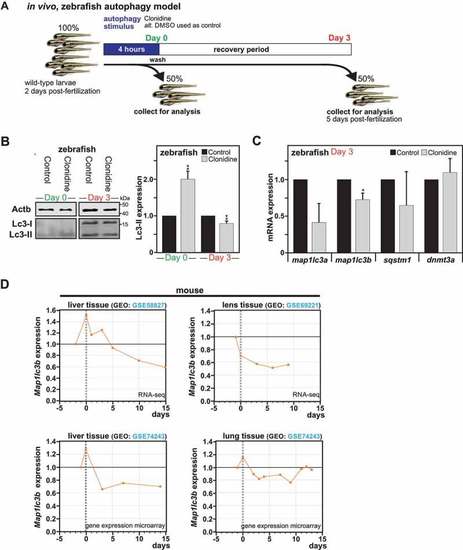
Impact of autophagy on long-term downregulation of MAP1LC3 expression |