- Title
-
Structural and functional conservation of non-lumenized lymphatic endothelial cells in the mammalian leptomeninges
- Authors
- Shibata-Germanos, S., Goodman, J.R., Grieg, A., Trivedi, C.A., Benson, B.C., Foti, S.C., Faro, A., Castellan, R.F.P., Correra, R.M., Barber, M., Ruhrberg, C., Weller, R.O., Lashley, T., Iliff, J.J., Hawkins, T.A., Rihel, J.
- Source
- Full text @ Acta Neuropathol.
|
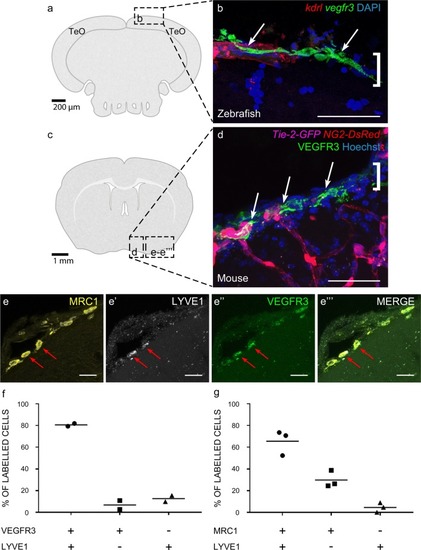
Cells with BLEC molecular markers are present within the mouse leptomeninges. |
|
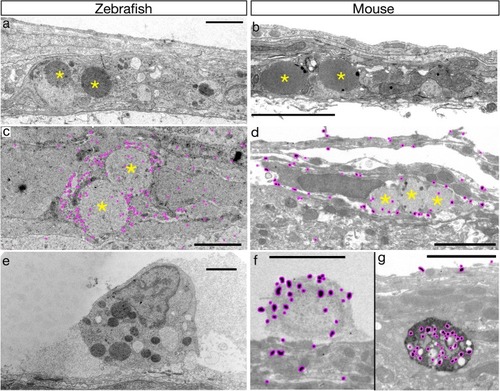
Ultrastructural properties of BLECs/LLECs are conserved in zebrafish and mice. |
|
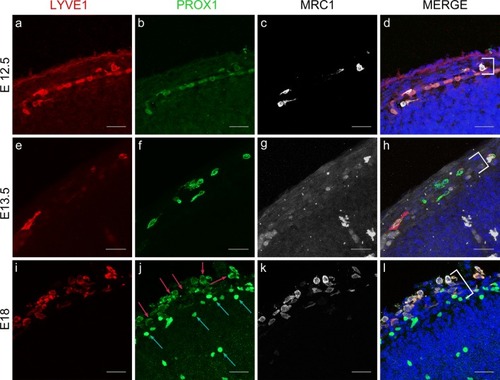
Cells expressing LLEC markers are present within embryonic mouse leptomeninges. |
|
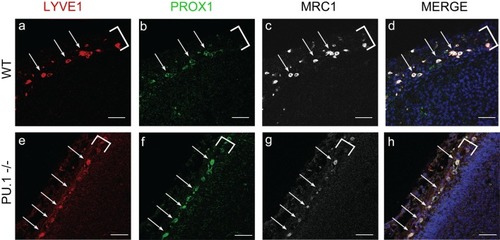
Mouse LLECs develop independent of the transcription factor PU.1. |
|
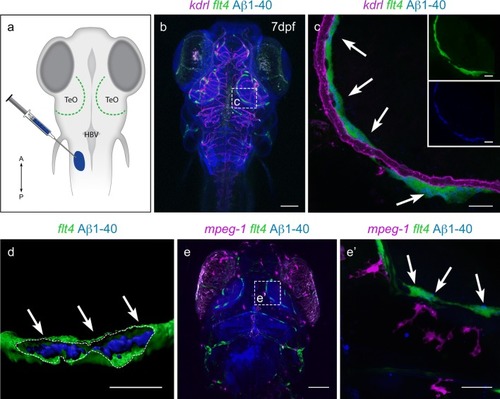
Zebrafish BLECs, but not macrophages, take up Aβ1-40. |
|
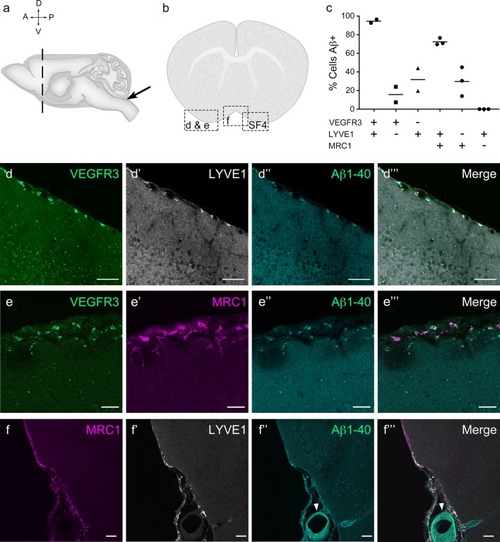
Mouse LLECs take up Aβ 1-40. |
|
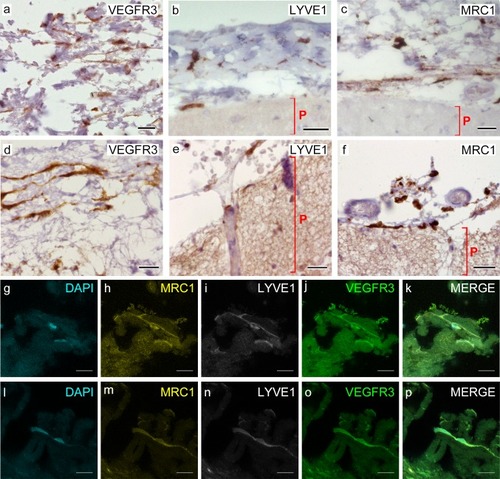
Cells of human meninges co-express LLEC markers. |