- Title
-
Fibrillarin is essential for S-phase progression and neuronal differentiation in zebrafish dorsal midbrain and retina
- Authors
- Bouffard, S., Dambroise, E., Brombin, A., Lempereur, S., Hatin, I., Simion, M., Corre, R., Bourrat, F., Joly, J.S., Jamen, F.
- Source
- Full text @ Dev. Biol.
|
fbl transcripts and Fbl protein are preferentially expressed in neural progenitors during zebrafish development. (A-E)In situ hybridization showing the progressive restriction of fbl expression during the development of zebrafish embryos (A)fbl is ubiquitously expressed at 6 hpf. (B). fbl expression begins to be restricted to highly proliferative regions (eyes, midbrain, and somites) during neurulation (C) At 24 hpf, fbl transcripts are abundant in the optic tectum (red arrows) and the retina. C1-C2: lateral views, C3: dorsal view (D-E) At 2 dpf (D) and 3 dpf (E), fbl is preferentially expressed in the neuroepithelial progenitors of the TMZe at the periphery of the OT (red arrows), and in the external edge of the ciliary marginal zone (CMZe) of the retina (green arrows). Additional expression can be detected in the digestive system D1-D2 and E1-E2: lateral views, D3 and E3: dorsal views. Scale bars: 100 µm. Anterior is to the left. (F) F1: Drawing of a 2dpf zebrafish embryo in dorsal view. The dark line corresponds to the sagittal section represented in F2. F2: Immunostaining showing Fbl protein on a sagittal section of a 2 dpf embryo. The Fbl protein, which is localized in the nucleoli, is present in all cells (yellow dots) but the punctate domains of expression are larger in TMZe neuroepithelial progenitors (surrounded by white dashed lines) than in other cells in the optic tectum (OT), cerebellum or torus semicircularis. (G) G1: Drawing of a 3dpf zebrafish embryo in dorsal view. The dark line corresponds to the transverse section represented in G2. G2: Immunostaining showing Fbl protein on a transvere section of a 3dpf embryo. The Fbl protein accumulates at the periphery of the retina where neuroepithelial cells are located (CMZe). Scale bars: 25 µm. Cb: cerebellum; CMZe: external edge of the ciliary marginal zone; OT: optic tectum; Ts: torus semicircularis;; TMZe: external edge of the tectal marginal zone. EXPRESSION / LABELING:
|
|
Gross morphological defects in fbl null mutants are mainly seen in the head and eyes and are rescued by injection of fbl mRNA. (A) RT-qPCR quantification of relative levels of fbl mRNA at the indicated developmental stages shows the absence of fbl expression in mutants as early as the 24 hpf stage. Purple: wild-type, gray: siblings, pink: mutants. Statistical analyses were performed on four biological samples per condition. p-value (Kruskal-Wallis test) polysomes: 0.02. n.s., non-significant. (B1-B6) Lateral views of live embryos with anterior to the left and dorsal up. (B1-B2)fblhi2581Tg mutants start to display phenotypic abnormalities as early as the 24 hpf stage with necrosis in the midbrain area, slightly smaller eyes and a thin yolk extension. (B3-B4) At 48 hpf, mutant embryos have smaller eyes and smaller head than their siblings. In addition, mutant embryos display less pigmentation, pericardiac edema, a larger and rounder yolk and a thinner yolk extension. (B5-B6) At 48 hpf, fbl mutant embryos are largely rescued by fbl mRNA. In injected mutants, midbrain and yolk extension look similar in size to the ones in siblings. The size of the eye is bigger than in not injected mutants although remaining slightly smaller than in siblings. In panels B1-B9, eyes, midbrain area and yolk extension are respectively indicated by arrowheads, arrows and asterisks. Scale bar = 200 µm. (C) Classes of phenotypes of fbl mRNA injected embryos from an fblhi2581Tg incross. A strong decrease in the number of injected embryos with mutant phenotype is observed indicating that the fbl mRNA rescues the mutation. EXPRESSION / LABELING:
PHENOTYPE:
|
|
The fblhi2581Tg mutation mostly affects midbrain structures from 24 hpf. (A) Volume rendering of the DiI-positive domains (gray) and surface rendering of a manual segmentation of the CNS (magenta) and eye (white) based on the DiI signal in 3dpf wild-type, fbl siblings and fblhi2581Tg mutant embryos. fbl mutant larvae display an apparent reduction of the CNS volume compared to their siblings or wild-type larvae Lateral views: anterior to the left, dorsal to the top. Dorsal view: anterior to the left, right to the top. (B) Quantification of mean CNS volume highlights a significant difference between fbl mutants, their siblings and wild-type larvae. Statistical analyses were performed on six samples per condition. p-value: .003 (Kruskal-Wallis test). (C) Quantification of mean eye volume highlights a significant difference between fbl mutants, their siblings and wild-type larvae. Statistical analyses were performed on the mean size of both eye of six samples per condition. p-value: .005 (Kruskal-Wallis test). Purple: wild-type, gray: fbl siblings, pink: fblhi2581Tg. Scale bar: 100 µm. Anterior is to the left. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
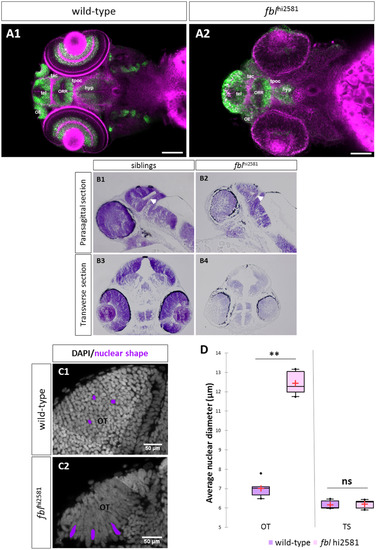
fbl mutants have specific midbrain and retina defects. (A) Horizontal optical sections of Elavl3 (marker of neural differentiation) immunolabelling and DiI staining in wild-type (A1) and mutant (A2) embryos at 3dpf. Pink: DiI labeling, Green: Elavl3 staining. Scale bars: 100 µm. Anterior is to the left. Most brain domains and axon tracts are present but have a disrupted organization in fbl mutant embryos. (B) Sagittal (A1-A2) and transverse (A3-A4) paraffin sections of wild-type (left) and fblhi2581Tg mutant (right) embryos at 2dpf. Histological analysis with cresyl violet staining revealed smaller tecta and acellular zones in the mutant embryos. The proliferation region (TMZe) is thicker in the mutant embryos (white arrow) (B2, B4) than in their siblings (B1, B3). Anterior is to the left. (C) Nuclear labeling (DAPI) in the optic tectum of 2dpf wild-type (C1) and mutant (C2) embryos at 2dpf, showing the larger nuclear diameter in the tectum of mutant larvae. Scale bar: 50 µm (D) Quantification of the nuclear diameter of wild-type (purple) and fblhi2581Tg (pink) 2dpf embryos. Nuclear diameters were measured with Fiji software. We measured the longest dimension of 50 nuclei on selected 2D images of the nuclei of 2 dpf embryos. Statistical analyses were performed on the mean diameters of nuclei from five mutant or wild-type embryos. p-value (Mann&Whitney test) OT: 0.008; p-value TS: 1.000. Hyp: hypothalamus, OE: olfactory epithelium, ORR: optic recess region OT: optic tectum, tac: tract of the anterior commissure, tel: telencephalon, tpoc: tract of the post-optic commissure, TS: torus semicircularis. PHENOTYPE:
|
|
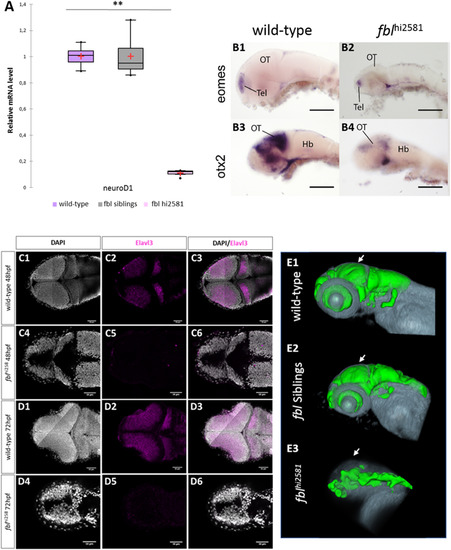
Neural specification and neural differentiation are impaired in fblhi2581Tg mutant embryos. (A) RT-qPCR quantification of the relative levels of neuroD1 mRNA. Purple: wild-type, gray: siblings, pink: mutants. (B) Expression patterns of eomes and otx2, two markers of neural specification, in 3 dpf wild-type (B1, B3) and fblhi2581Tg mutant embryos (B2, B4). The expression of eomes(B1-B2), a marker of forebrain specification, was similar in wild-type and mutant embryos, whereas that of otx2(B3-B4), a marker of midbrain specification, is absent in fblhi2581Tg mutant embryos. Scale bars: 50 µm. Hb: hindbrain, OT: optic tectum, Tel: telencephalon. (C) Horizontal optical sections of elavl3 (marker of neural differentiation) labeling in 2dpf wild-type (C1-C3) and mutant (C4-C6) embryos and in 3dpf wild-type (D1-D3) and mutant (D4-D6) embryos. Gray: DAPI staining, pink: Elavl3 staining. Scale bars: 50 µm. Anterior is to the left. (E) Volume rendering of the DiI-positive domains (gray) and surface rendering of a manual segmentation of the Elavl3-positive (green) domains in 3dpf wild-type (E1), fbl siblings (E2) and fblhi2381 mutant embryos (E3). For lateral views, anterior to the left and dorsal to the top. White arrows point out to the midbrain. Neural differentiation is specifically impaired dorsally in fbl mutant. EXPRESSION / LABELING:
PHENOTYPE:
|
|
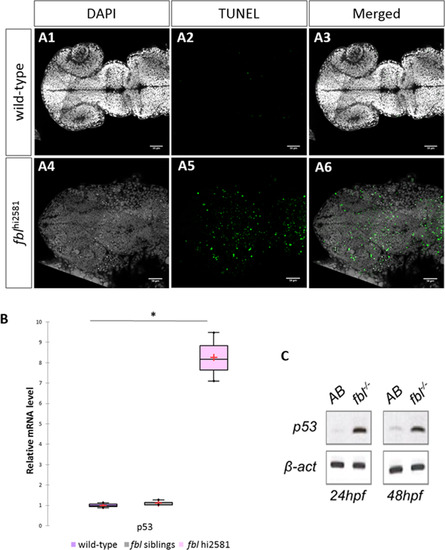
Massive p53-dependent apoptosis in the fbl mutant. (A) Horizontal optic sections of TUNEL labeling at 24 hpf in wild-type (A1-A3) and mutant (A4-A6) embryos. Gray: DAPI staining, Green: TUNEL staining. Scale bar: 50 µm. Anterior is to the left. (B) RT-qPCR quantification of relative levels of tp53 mRNA at 72 hpf shows a strong increase in tp53 expression in mutants. Purple: wild-type, gray: siblings, pink: mutants. Statistical analyses were performed on biological triplicates, p-value (Kruskal-Wallis test): 0.049. (C) RT-PCR for tp53 in 24 hpf and 48 hpf mutant embryos showing a large increase in tp53 expression. Anterior is to the left. EXPRESSION / LABELING:
PHENOTYPE:
|
|
In fbl mutants, the S-phase of the cell cycle is affected. (A) PCNA staining in 2dpf wild type (A1) and mutant (A2). In wild-type larvae, proliferative cells were restricted to the periphery of each tectal lobes. In fblhi2581Tg PCNA positive cells were observed in almost all tectal cells. (B) EdU incorporation experiments in 2dpf wild type (A1) and mutant (A2) embryos after two hour pulse. In wild-type embryos, EdU-positive cells are restricted to the periphery of the OT while in the fblhi2581Tg mutant embryos EdU-positive cells are spread all over the structure. (C) EdU-positive cells quantification in wild-type (purple) and mutant (pink) embryos at 2dpf. Statistical analyzes have been performed on four samples per conditions, p-value (Mann&Whitney test): 1.00. (D) pH3 staining in 2dpf wild types (E1) and mutants (E2) embryos. Similar abnormal patterns in mutants as for EdU incorporation experiments. (E) pH3-positive cells quantification in wild-type (purple) and mutant (pink) embryos at 2dpf. Statistical analyzes have been performed on four samples per conditions, p-value (Mann&Whitney test): 0.53 (E) embryos. Gray: DAPI staining, Pink: EdU, pH3 or PCNA staining. Scale bar: 50 µm. Anterior is to the left. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|
Reprinted from Developmental Biology, 437(1), Bouffard, S., Dambroise, E., Brombin, A., Lempereur, S., Hatin, I., Simion, M., Corre, R., Bourrat, F., Joly, J.S., Jamen, F., Fibrillarin is essential for S-phase progression and neuronal differentiation in zebrafish dorsal midbrain and retina, 1-16, Copyright (2018) with permission from Elsevier. Full text @ Dev. Biol.