- Title
-
A histone demethylase is necessary for regeneration in zebrafish
- Authors
- Stewart, S., Tsun, Z.Y., and Izpisúa Belmonte, J.C.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
me3K27 H3 demethylases in caudal fin. (A) Global levels of me3K27 H3 remain intact during regeneration. Immunohistochemistry of longitudinal sections of 48 hpa regenerating caudal fins. From left to right, me3K27 H3 antibody; me3K27 H3 antibody with peptide competition; msxB; PCNA; p63. The right-most panel is a non-regenerating caudal fin stained for me3K27 H3. Antibody signal (brown) and hematoxylin counterstain (blue) are shown. (B–E) Expression of putative me3K27 H3 demethylases in regenerating caudal fins. Whole mount in situ hybridization of regenerating caudal fins at 48 h post amputation (B) kdm6a.1, (C) kdm6a.2, (D) kdm6b.2, and (E) kdm6b.1. Longitudinal sections of the fins in (B–E) reveal unique expression patterns and are shown in the right panels. The epidermis (e) and mesenchyme (m) are indicated in each panel. (F) Upregulation of kdm6b.1 during regeneration. The expression of putative me3K27 H3 demethylases was examined in non-regenerating and regenerating (48 hpa) adult caudal fins by qPCR and normalized to Ribosomal Protein L18 expression. Data are expressed as average expression levels relative to non-regenerating fins from two different cohorts (12 animals each) and error bars represent the deviation from the means derived from each cohort. Similar results were obtained in several independent experiments. (G) kdm6b.1 expression in larvae caudal fin at 24 hpa [72 h post fertilization (hpf)] by in situ hybridization. (H) Kdm6b.1 demethylates me3K27 H3. myc-tagged Kdm6b.1 was immunoprecipitated from transfected 293T cells and subjected to a histone demethylase assay, followed by immunoblotting. Some panels contain blots that were stripped and re-probed with different antibodies. EXPRESSION / LABELING:
|
|
Kdm6b.1 is needed for fin regeneration. Analysis of regeneration in control versus Kdm6b.1 MO1, MO3, or MO4 injected animals was followed over time in individual animals. Representative examples of a control (upper panels) and Kdm6b.1 MO (lower panels) phenotypes are shown before amputation, immediately after amputation, and 24 and 48 h after amputation. The amputation planes are similar to that shown above and the same animal is shown before and after amputation up to 48 h. PHENOTYPE:
|
|
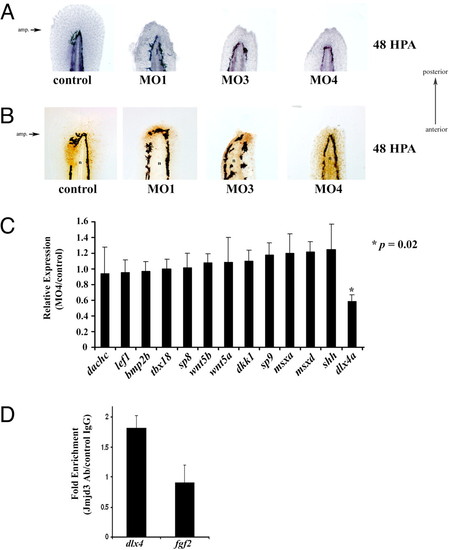
Identification of dlx4a as putative target of Kdm6b.1. (A) Control or Kdm6b.1 morpholino injected animals were amputated and allowed to regenerate for 48 h at which point they were fixed and stained with hematoxylin. The amputation planes are similar to that shown above. (B) Proliferating cells in control and Kdm6b.1 morphants. Control (left panel) or Kdm6b.1 MO (right panels) for 48 h, at which point they were stained with PCNA antibodies (PCNA+ cells are stained brown). n, notochord. The amputation planes are similar to that shown above. (C) dlx4a is downregulated in Kdm6b.1 MO animals. The expression levels of various candidate genes were tested in Kdm6 b.1 MO4 and control animals at 96 hpf (24 hpa) and normalized to Ribosomal Protein L18 expression. Data are expressed as average expression levels in the morphants relative to control animals. Error bars, the standard deviation and an asterisk (*) indicates a P value of 0.02 (n = 6 animals for each). (D) Jmjd3 associates with the dlx4 promoter in developing limb buds. Limb buds from approximately 20 pups were harvested at day e10.5 and processed for ChIP with control or Jmjd3 antibodies, followed by qPCR using primers at the transcription start sites of dlx4 or fgf2. Shown are the data from one experiment; error bars indicate the deviation from the mean duplicate ChIPs. Similar data were obtained in another experiment from different limb buds. PHENOTYPE:
|
|
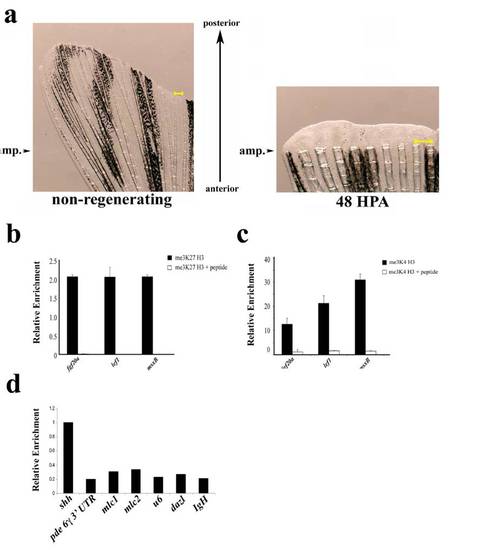
(a) Examples of uncut and regenerating fins. Arrowheads indicate the site of amputation on an un-amputated fin (Left) or a fin at 48 h post amputation (Right). Tissue distal to these arrowheads was used for ChIP or gene expression analysis. The regenerating fin is magnified to allow visualization of the newly formed regenerating tissue and a scale bar (yellow) is shown in each panel. (b and c) Specificity of methyl histone ChIPs. me3K27 H3 ChIP (b) or me3K4 H3 ChIP (c) were performed from non-regenerating chromatin with the indicated antibodies that had been preincubated with either BSA (1 mg/mL) or peptide antigens (10χmolar excess). Preincubation of antibody with peptide significantly reduces amount of eluted material relative to control. (d) Examples of genomic regions of relatively high and low levels of me3K4 H3. me3K4 H3 ChIP was performed as above (without peptide competition) and eluates were examined by real-time PCR with the indicated primer sets. Data are expressed relative to the ChIP of the transcription start site of shah to highlight the significantly lower levels of me3K4 H3 at the other genomic positions. Primers used were: 3′ UTR of phosphodiesterase 6 gamma; cardiac myosin light chain 1 (mlc1) promoter; cardiac myosin light chain 2 (mlc2) promoter; u6 small RNA promoter; dazl promoter; IgH diversity region intronic sequence (IgH). |
|
(a) Zebrafish proximal caudal fin tissue. The yellow highlighted region roughly indicates the region used for chromatin extraction and ChIP analysis. (b and c) Bivalent chromatin in non-regenerating zebrafish proximal caudal fin tissue. (b) Sequential ChIP first performed with me3K4 H3 followed by control or me3K27 H3 antibody. (c) Sequential ChIP first performed with me3K27 H3 followed by control or me3K4 H3antibody. Eluates from the second ChIP were examined by qPCR using primers near the transcription start sites of the indicated genes. Shown is the average of two independent sequential ChIPs; error bars indicate deviation from the mean from two cohorts. |
|
Whole mount and longitudinal sections of in situ hybridization on 48 hpa regenerating caudal fins (a–h) a, eed b, ezh2 c, suz12 d, fgf20a e, lef1 f, msxb. Longitudinal sections from the same fin are shown in the Right. The epidermis (e) and mesenchyme (m) are indicated in each panel. (g) Upregulation of Polycomb genes during regeneration. The expression of eed, ezh1, ezh2, and suz12 were examined in non-regenerating and regenerating (48 hpa) caudal by qPCR from caudal fin cDNA from two cohorts of animals. Expression levels were first normalized to ribosomal protein L18 expression. The fold change was calculated by dividing regenerating levels by non-regenerating expression levels for each gene examined. Error bars indicate the deviation from the mean. Similar data were obtained in at least three independent experiments. |
|
(a) DZNEP inhibits regeneration. DZNEP (neoplanocin A) is a direct inhibitor of S-adenosyl methionine homocysteine hydrolase, an enzyme with roles in both epigenetic silencing and cellular senescence (43–47). Caudal fins of adult animals were amputated and animals were allowed to regenerate in the presence or absence of DZNEP (15 μM) for 6 days at which point they were photographed. Shown is a typical animal (n > 10). (b) Caudal fins from control or DZNEP (15μM) treated animals at 48 hpa were sectioned and stained with the indicated antibodies. The epidermis (e) and mesenchyme (m) are indicated in each panel. (c) DZNEP does not alter global levels of histone methylation. Animals were treated as above (15μM DZNEP) and caudal fins were harvested, homogenized and boiled in SDS buffer. Extracts were subjected to SDS/PAGE and immunoblot analysis with the indicated antibodies. Some panels were stripped and re-probed with different antibodies. (d) DZENP treatment upregulates msxe and tbx18 gene expression. qPCR was used to examine differences in gene expression between DMSOand DZNEP (15μM) treated animals at 48 hpa. Data represent relative expression changes of genes (treated/untreated) at 48 hpa normalized to ribosomal protein L 18 gene expression. Error bars indicate standard deviation from the mean (n = 3). An asterisk (*) indicates a P value of less than 0.05 (comparing cDNA derived from DMSO versus DZNEP treated animals) which were calculated using a two-tailed paired t-test. (e) DZENP reduces me3K27 H3 levels at the promoters of msxe and tbx18. Extracts from DMSO- and DZNEP-treated animals (15 μM) at 48 hpa were subjected to ChIP with me3K27 H3 antibodies followed by qPCR with the primers at the transcription start sites at the indicated loci. Data are expressed as DZEP animal ChIP/control animal ChIP. |
|
Whole mount in situ hybridization of Polycomb PRC1 components during caudal fin regeneration at 48 hpa (a–f). Longitudinal sections of caudal fins demonstrating PRC1 components expression during regeneration at 48 hpa (g–k). Ubiquitous presence of Ub-H2AK119 in caudal fin detected by immunohistochemistry (l). IHC was performed on PFA fixed longitudinal sections of fins at 48 hpa. The epidermis (e) and mesenchyme (m) are indicated in each panel. Antibody signal (brown) and hematoxylin counterstain (blue) are shown. |
|
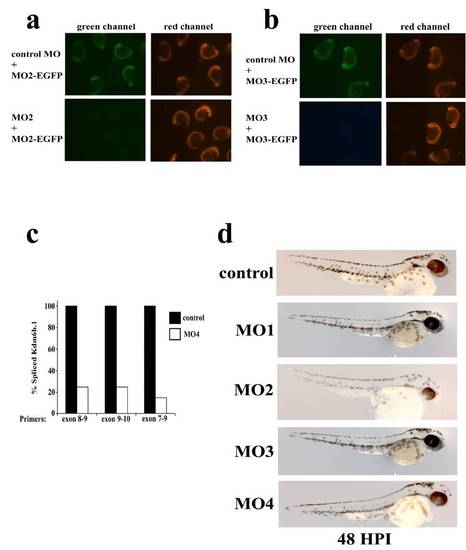
Morpholino control experiments. (a and b) To demonstrate efficacy of the morpholinos, the target sequence for MO2 and MO3 was cloned upstream of EGFP in pCS2 vector. Animals were injected with RNA transcribed from these vectors in the presence of mRFP RNA and the indicated morpholino. Twenty-four hpi animals were illuminated under epifluorescence and photographed. Exactly the same exposure times were used in each of the four panels shown. MO1 is a FITC labeled translation blocking morpholino which prevented us from performing the heterologous reporter assay as was done for MO2 and MO3. (c) In the case of MO4, real-time PCR was performed on cDNA derived from control or MO4 injected animals at 24 hpi. Primers were designed to amplify regions of the correctly spliced Kdm6b.1 mRNA around exon 8 which is the predicted target of MO4. (d) Animals injected at the one-cell stage with the indicated morpholino were photographed at 48 hpi. At these doses of morpholinos a large number of animals develop normally and were used for regeneration studies. |
|
Kdm6b.1 MO2 inhibits regeneration. (a and b). Embryos were injected at the one-cell stage with approximately 1 nL control or Kdm6b.1 MO2 morpholinos (250 μM needle concentration). After 48 h, animals were sedated and caudal fin primordial amputated distal to the notochord. Animals were harvested 48 additional h later and fixed and photographed or stained with hematoxylin or PCNA antibodies as indicated (b). (c) Regeneration is abrogated up to 96 hpa in Km6b.1 knockdown animals compared to control animals. |