- Title
-
DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45
- Authors
- Rai, K., Huggins, I.J., James, S.R., Karpf, A.R., Jones, D.A., and Cairns, B.R.
- Source
- Full text @ Cell
|
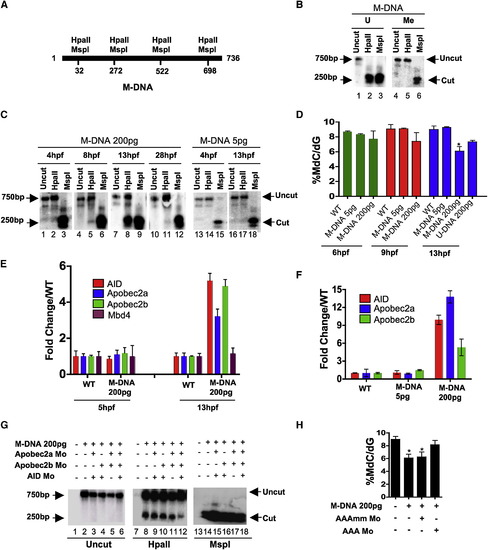
Involvement of Cytidine Deaminases in DNA Demethylation in Zebrafish (A) Schematic of the M-DNA (Methylated DNA) fragment injected into fertilized embryos at the single-cell stage. The four HpaII/MspI sites are indicated. HpaII-resistant (methylated CmeCGG) and HpaII-cleaved species (unmethylated) run at ∼750 bp and ∼250 bp, respectively (B). (B) M-DNA methylation status was assessed by HpaII susceptibility, with cutting observed on unmethylated (U) (lane 2) but not methylated (Me) DNA (lane 2). MspI-digested (lanes 3 and 6) and uncut DNA (lanes 1 and 4) served as controls. Fragments were detected by Southern blot analysis probed with full-length M-DNA probe. (C) M-DNA methylation status during development. Total DNA was isolated at the time points shown from embryos injected at the single-cell stage with M-DNA (5 pg or 200 pg), and treated as in (B). M-DNA induced demethylation peaks at ∼13 hpf (Compare lanes 2, 5, 8, and 11). (D) LC-MS quantitation of 5-meC content of the bulk genome (normalized to total deoxyguanosine content) in genomic DNA isolated from embryos (hpf indicated) injected with methylated M-DNA or unmethylated U-DNA at the single-cell stage. (E and F) qRT-PCR determinations from embryos injected with M-DNA (E), and at different fragment concentrations (F). (G and H) Methylation status of M-DNA assessed by HpaII digestion and Southern blotting (G), or LC-MS quantitation of total 5-MeC (H) in total genomic DNA isolated from embryos at 13 hpf, injected at the single-cell stage with M-DNA (200 pg) and morpholinos as indicated. Lanes 1, 7, and 13 correspond to wild-type sample. AAAmm refers to a set of three control morpholinos against AID (4 pg), Apobec2a (4 pg), and Apobec2b (2 pg) (AAA), which each contain five mismatched (mm) bases (of 25 total to prevent binding) relative to the efficacious morpholino (same amount as controls). For HpaII/MspI susceptibility, one representative of at least three biological repeats is shown. LC-MS measurements; two biological replicates. Asterisks (*) depict statistical significance (p < 0.05). Error bars: +/- one standard deviation. |
|
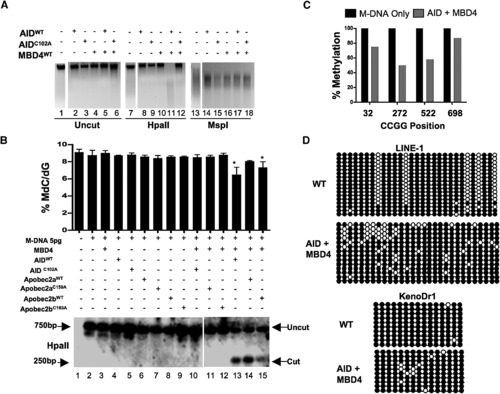
Overexpression of a Deaminase/Glycosylase Pair Elicits DNA Demethylation (A–C) Methylation status assessed by HpaII digestion of total genomic DNA (A), LC-MS quantitation ([B] upper panel), HpaII digestion of M-DNA (Southern analysis) ([B] lower panel), and bisulphite sequencing of M-DNA (C). Lanes 1, 7, and 13 in (A) and lane 1 in (B) correspond to wild-type sample. For (B), M-DNA was injected at 5 pg, below the threshold level for eliciting demethylation on its own (See Figures 1C and 1D). For (C), twenty clones were subjected to bisulphite sequencing, and the methylation status of each HpaII/MspI (CCGG) site reported as a percentage of total sites tested. (D) Repeat elements from DNA isolated from embryos (13 hpf) injected at the single-cell stage with RNA encoding wild-type AID, along with MBD4 wild-type mRNA. For each experiment, one representative of at least three biological repeats is shown except in LC-MS measurement where graph is prepared from values of two biological replicates. Asterisks (*) depict statistical significance (p < 0.05). Error bars are ± one standard deviation. |
|
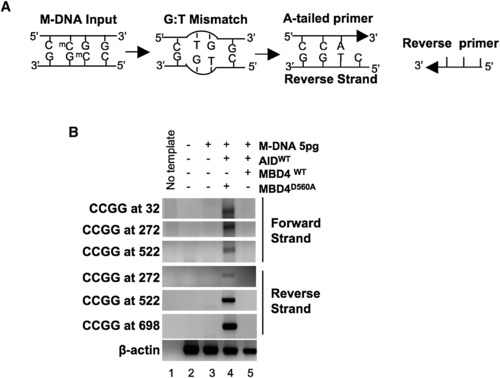
A PCR Strategy to Detect a G:T Intermediate (A) Schematic of the PCR reaction for thymine (CmeCGG > CTGG) detection at M-DNA HpaII/MspI sites using an A-tailed primer (only 3 of the ∼22 bases shown) with an adenosine at the 3′ end. (B) Detection of a G:T mismatch on M-DNA by PCR. M-DNA, AID mRNA, and RNA encoding either wild-type or catalytically inactive hMbd4 (D560A) was injected at the single-cell stage and assessed at 13 hpf. |
|
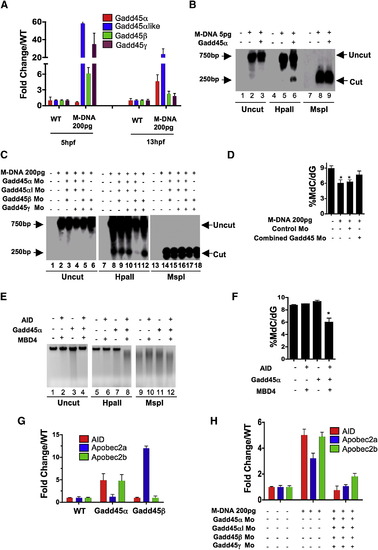
Gadd45 Proteins Promote Demethylation and Selectively Upregulate Deaminases (A) Gadd45 family members are upregulated by M-DNA, assessed by RT-PCR. (B) Gadd45α induces moderate demethylation of M-DNA as detected by HpaII digestion and subsequent Southern blotting. M-DNA is injected at 5 pg which does not induce demethylation on its own. Lanes 1, 4, and 7 correspond to wild-type sample. (C and D) Lowering the levels of Gadd45 family members via morpholino injection attenuates demethylation. Demethylation of 5-methylcytosine as assessed by HpaII digestion and Southern blotting (C), or LC-MS quantitation of 5-meC (D) in total genomic DNA isolated from 13 hpf old embryos injected at the single-cell stage with M-DNA alone (200 pgs) or along with morpholinos as shown. Lanes 1, 7 and 13 correspond to wild-type sample. Combined Gadd45 Mo refers to the combination of morpholinos to all four Gadd45 family members tested (α,α -like, β, and γ; 2 pg each). (E and F) Synergy among AID, hMbd4, and Gadd45 for demethylation. Methylation status of total genomic DNA from embryos (13 hpf) was assessed by HpaII susceptibility (E) or by LC-MS quantitation of global 5-methylcytosine levels (F) injected at single-cell stage with mRNAs encoding factors as indicated. Lanes 1, 5, and 9 correspond to wild-type sample. Note: AID, MBD4, and Gadd45α were injected at subthreshold amounts (at 25 pgs each), levels which are not sufficient to induce demethylation alone. (G and H) Quantitative RT-PCR for deaminase family members (assessed at 13 hpf) injected at single-cell stage with mRNA encoding Gadd45α or Gadd45β (F) and M-DNA (200 pg) alone or with morpholinos as shown (G). For each experiment, one representative of at least three biological repeats is shown except in LC-MS measurement where graph is prepared from values of two biological replicates. Asterisks (*) depict statistical significance (p < 0.05). Error bars: are ± one standard deviation. |
|
Methylated plasmid induces demethylation in zebrafish |
|
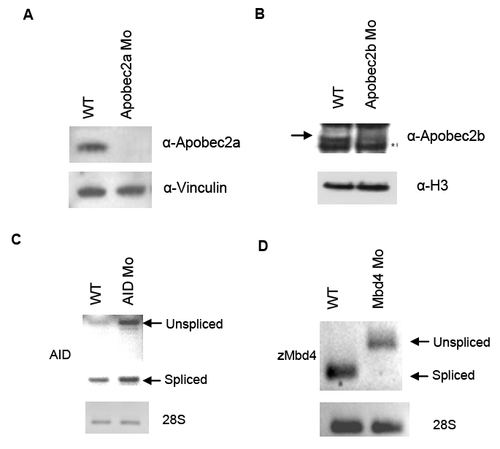
Efficiency of morpholino knockdown of deaminase proteins and MBD4 |
|
AID morphants, zMbd4 morphants, and Gadd45α morphants show neurogenesis defects |
|
Protein expression levels of catalytic mutant derivatives |
|
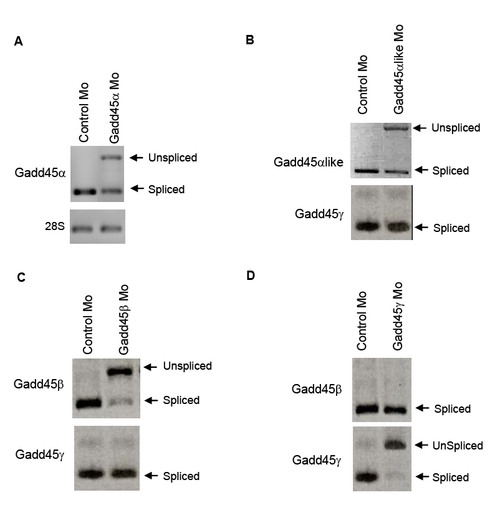
Morpholino knockdown efficacy of Gadd45 family members |
|
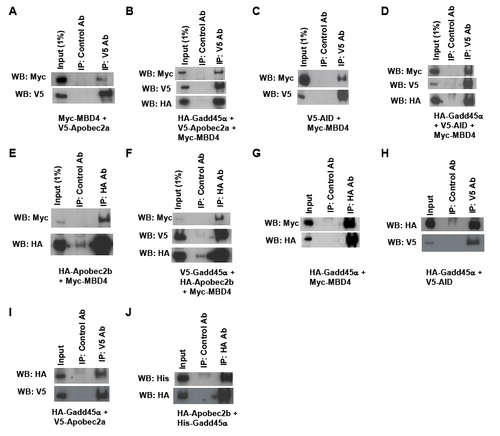
Physical interactions between AID and Mbd4 and enhancement by Gadd45α |
Reprinted from Cell, 135(7), Rai, K., Huggins, I.J., James, S.R., Karpf, A.R., Jones, D.A., and Cairns, B.R., DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45, 1201-1212, Copyright (2008) with permission from Elsevier. Full text @ Cell