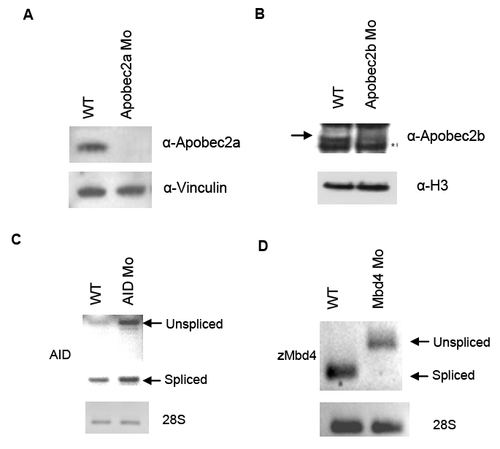
Fig. S3
|
Efficiency of morpholino knockdown of deaminase proteins and MBD4 |
Reprinted from Cell, 135(7), Rai, K., Huggins, I.J., James, S.R., Karpf, A.R., Jones, D.A., and Cairns, B.R., DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45, 1201-1212, Copyright (2008) with permission from Elsevier. Full text @ Cell