- Title
-
Key sequence features of CRISPR RNA for dual-guide CRISPR-Cas9 ribonucleoprotein complexes assembled with wild-type or HiFi Cas9
- Authors
- Okada, K., Aoki, K., Tabei, T., Sugio, K., Imai, K., Bonkohara, Y., Kamachi, Y.
- Source
- Full text @ Nucleic Acids Res.
|
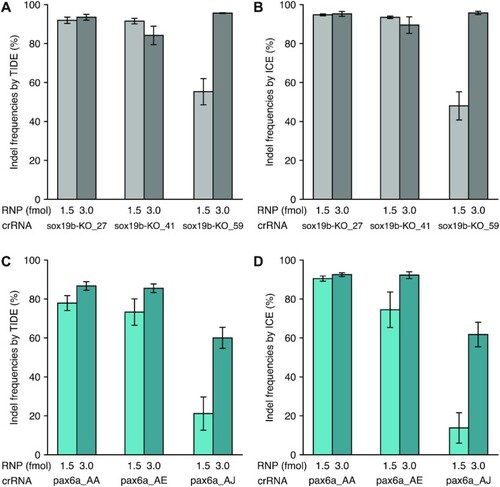
Establishing adequate quantities of CRISPR-Cas9 RNP complexes for crRNA evaluation. ( |
|
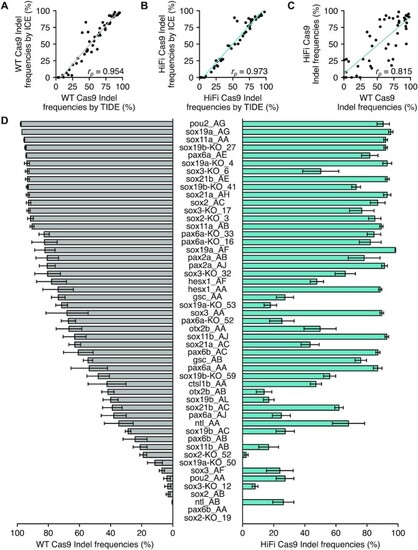
Cleavage efficiencies of the 51 crRNAs in the dgRNA RNP complex assembled with WT or HiFi Cas9. ( |
|
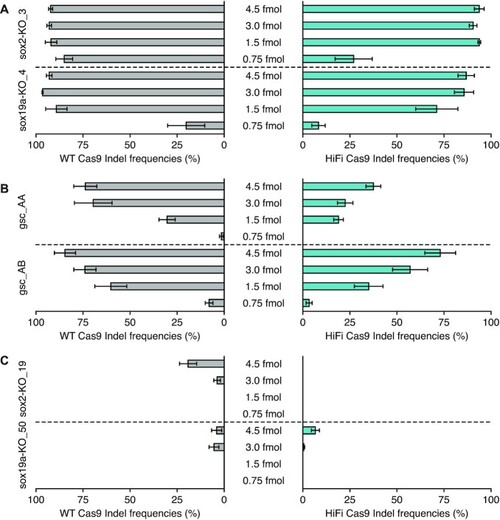
Dose-dependent cleavage efficiencies of selected crRNAs from low, medium, and high activity groups. Two crRNAs each from high ( |
|
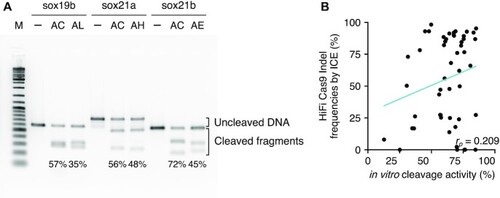
Assessment of cleavage efficiency of crRNA in an |
|
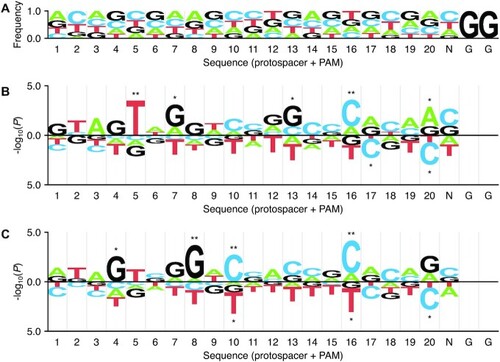
Position-specific mononucleotide features determined with kpLogo. ( |
|
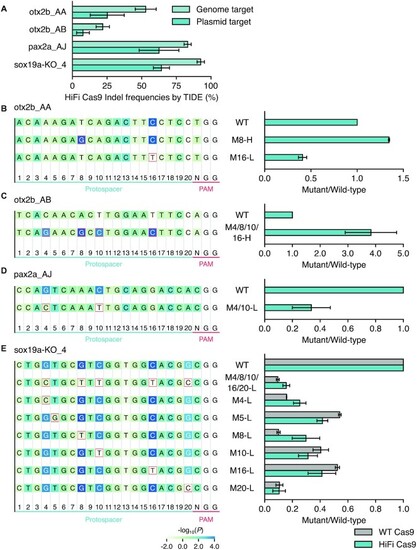
Experimental validation of mononucleotide features associated with crRNA efficiency. ( |
|
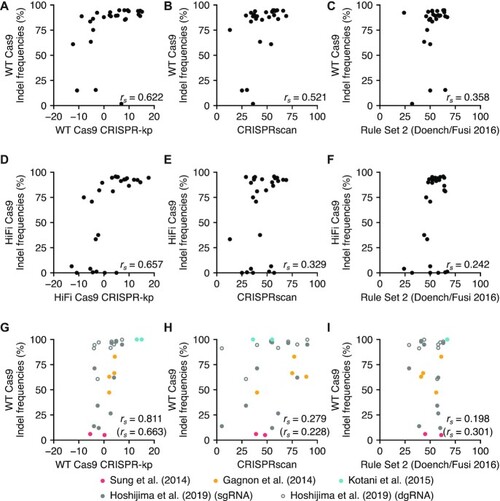
Predictive power of current CRISPR-Cas9 gRNA design tools for dgRNA/Cas9 RNPs. The heat map shows Spearman rank correlation coefficients ( |
|
Prediction of gRNA efficiency by CRISPR-kp, position-specific mononucleotide feature-based scoring. ( |
|
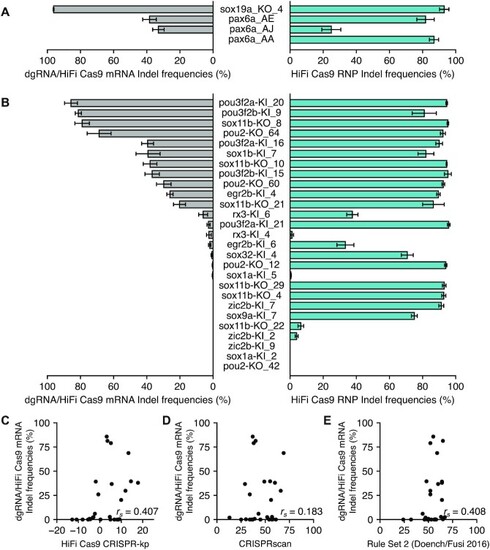
Comparison of dgRNA efficiency between injection of pre-assembled RNP and a dgRNA/Cas9 mRNA mixture. ( |