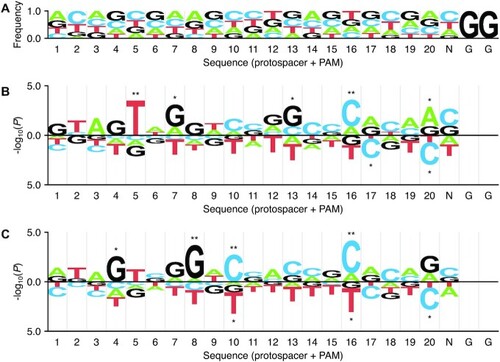
Figure 5.
- ID
- ZDB-FIG-220323-9
- Publication
- Okada et al., 2022 - Key sequence features of CRISPR RNA for dual-guide CRISPR-Cas9 ribonucleoprotein complexes assembled with wild-type or HiFi Cas9
- Other Figures
- All Figure Page
- Back to All Figure Page
|
Position-specific mononucleotide features determined with kpLogo. ( |