- Title
-
miR-128a Acts as a Regulator in Cardiac Development by Modulating Differentiation of Cardiac Progenitor Cell Populations
- Authors
- Hoelscher, S.C., Stich, T., Diehm, A., Lahm, H., Dreßen, M., Zhang, Z., Neb, I., Aherrahrou, Z., Erdmann, J., Schunkert, H., Santamaria, G., Cuda, G., Gilsbach, R., Hein, L., Lange, R., Hassel, D., Krane, M., Doppler, S.A.
- Source
- Full text @ Int. J. Mol. Sci.
|
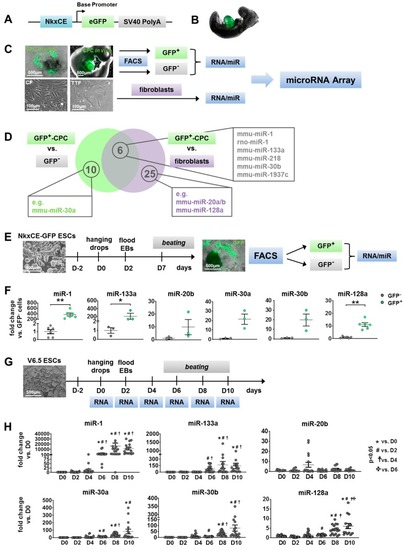
Identification of candidate microRNAs (miRs) during cardiac development. ( |
|
Evaluation of candidate miR function during zebrafish development. ( EXPRESSION / LABELING:
|
|
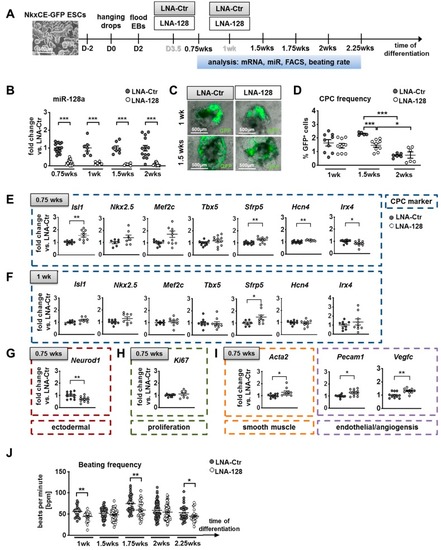
Knockdown of miR-128a during in vitro differentiation of murine NkxCE-GFP embryonic stem cells (ESCs). ( |
|
Knockdown of miR-128a during in vitro differentiation of murine Isl1-Cre/ROSA26mTmG induced pluripotent stem cells (iPSCs) (iITG-iPSCs). ( |
|
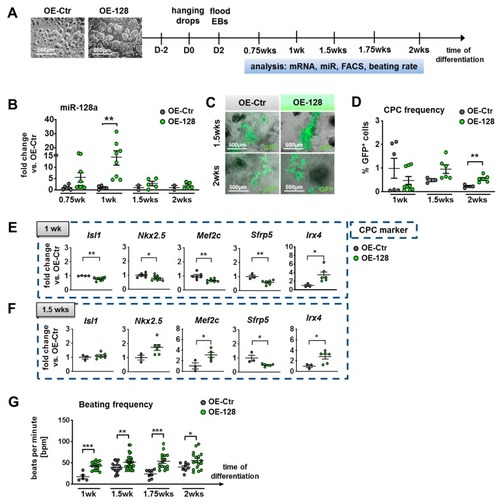
Overexpression (OE) of miR-128a during in vitro differentiation of murine NkxCE-GFP OE-128 and OE-Ctr ESCs. ( |
|
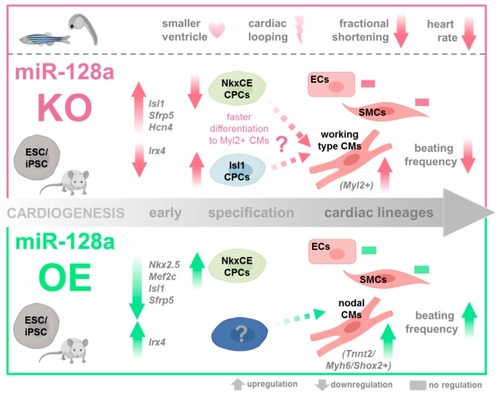
Role of miR-128a in cardiac development. During murine in vitro differentiation, LNA-mediated miR-128a knockdown in differentiating ES/iPSCs (1) increased cardiac transcription factors such as |