- Title
-
Global Transcriptomic Analysis of Zebrafish Glucagon Receptor Mutant Reveals Its Regulated Metabolic Network
- Authors
- Kang, Q., Hu, M., Jia, J., Bai, X., Liu, C., Wu, Z., Chen, W., Li, M.
- Source
- Full text @ Int. J. Mol. Sci.
|
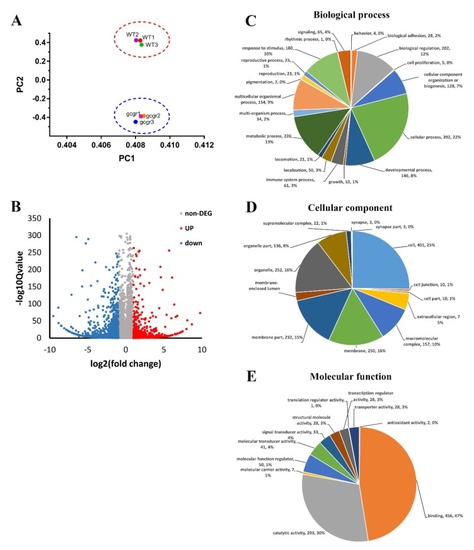
RNA-seq (RNA sequencing) analysis of glucagon receptor ( |
|
PHENOTYPE:
|
|
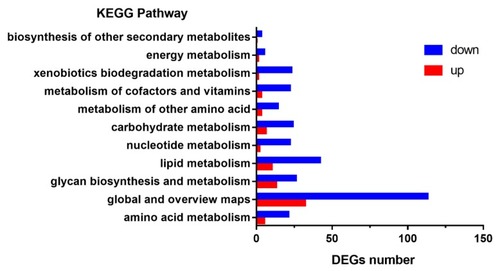
Kyoto encyclopedia of genes and genomes (KEGG) enrichment analysis of DEGs in metabolism pathways. The y-axis indicates pathways and the x-axis indicates the number of DEGs. The red bar shows the upregulated genes and the blue bar shows the downregulated genes. |
|
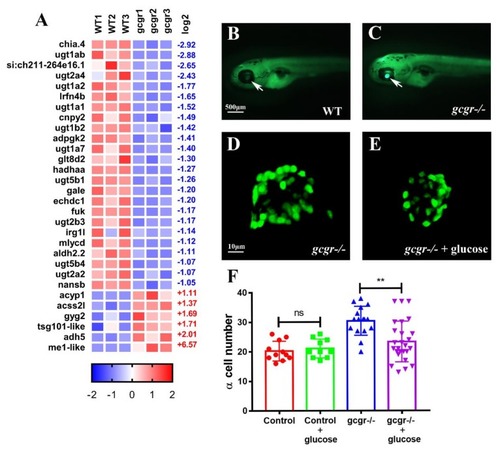
GCGR regulate lipid metabolism in zebrafish larva. ( PHENOTYPE:
|
|
GCGR regulates carbohydrate metabolism in zebrafish larvae. ( |
|
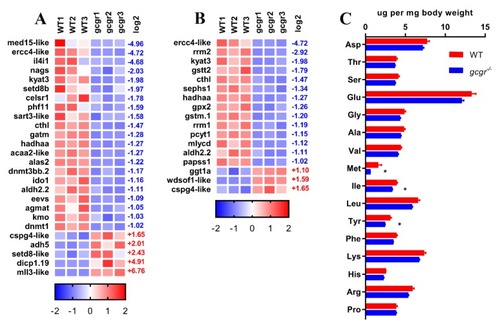
GCGR regulates amino acid metabolism in zebrafish larvae. ( PHENOTYPE:
|
|
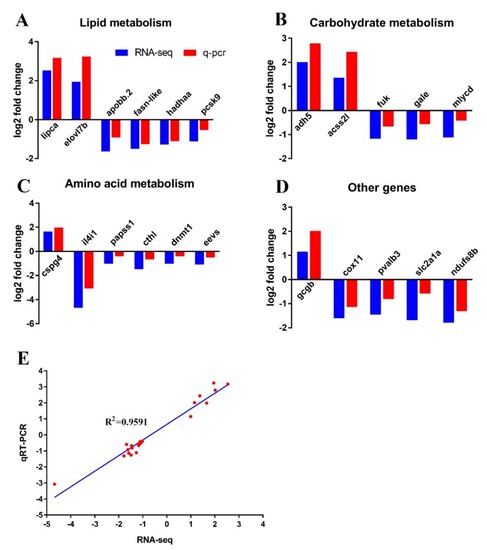
Validation of RNA-seq data using qPCR. ( |