Fig. 2
- ID
- ZDB-FIG-250805-54
- Publication
- Légaré et al., 2025 - Structural and genetic determinants of zebrafish functional brain networks
- Other Figures
- All Figure Page
- Back to All Figure Page
|
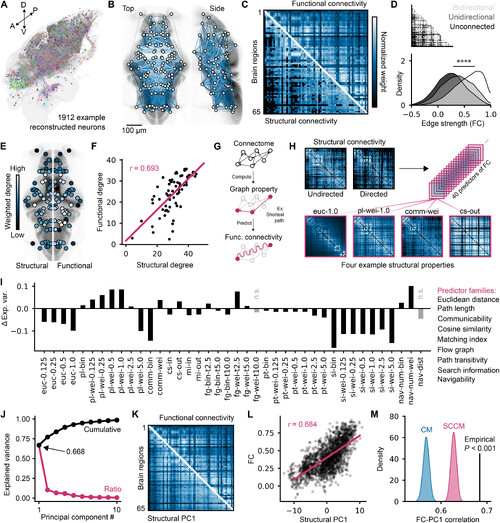
Structure- function coupling of zebrafish brain networks. (A) 3d rendering of 1912 reconstructed neurons generated on mapZebrain.org. (B) Mesoscopic Scoverlaid on brain anatomy; edge weights are color mapped with the next panel, and bottom quartile edges are not displayed. (C) Undirected Sc matrix compared withFc; matrices are arbitrarily scaled for visualization. (D) distributions of individually sampled Fc values from edges that have either bidirectional, unidirectional, or no un-derlying structural connections (one-way AnOvA, F = 5048, P = 0). (E) Functional and structural node degrees with arbitrary rescaled values for visualization; structuraldegrees are displayed on the left hemisphere, and functional degrees are displayed on the right. (F) Linear regression between structural and functional degrees (Pearson’sr = 0.693). (G) depiction of the Fc modeling approach used in following panels; properties of the structural network, such as the shortest path between two nodes, areused to predict Fc. (H) Four example matrices from an array of 40 graph properties used as predictors of Fc in this study. (I) Fc variance explained by each of the predictors,subtracted by the variance explained by Sc; predictor families are indicated on the right, in the same order as the bar chart labels. n.s., not significant. (J) Relative (red) andcumulative (black) variance explained by each Pc derived from the set of predictors. (K) Structural Pc1 compared to Fc; matrices are arbitrarily scaled for visualization.(L) Linear regression between upper triangle values of structural Pc1 and Fc (Pearson’s r = 0.684). (M) null distributions of structural Pc1 correlations derived from null Scmatrices (1000 matrices per distribution, empirical P < 0.001). |