Fig 4
- ID
- ZDB-FIG-250505-109
- Publication
- Dogra et al., 2025 - Modulation of NMDA receptor signaling and zinc chelation prevent seizure-like events in a zebrafish model of SLC13A5 epilepsy
- Other Figures
- All Figure Page
- Back to All Figure Page
|
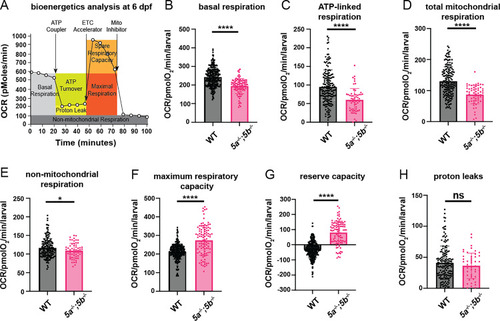
Metabolic health analysis in (A) Schematic representation of how the Seahorse bioanalyzer displays mitochondrial bioenergetics being regulated by pharmacological inhibitors. (B) Quantification of basal respiration at 6 dpf. |
| Fish: | |
|---|---|
| Observed In: | |
| Stage: | Day 6 |