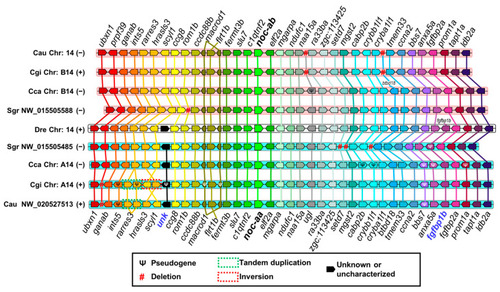
Synteny analysis of noc-a paralogues in Danio rerio (Dre), Sinocyclocheilus graham (Sgr), Cyprinus carpio (Cca), Carassius gibelio (Cgi), and Carassius auratus (Cau). Black bolded names indicate nocturnin paralogues in the center of synteny. Chromosomes are represented with a white background in Danio rerio (as the reference species) and a cyan or pink background in the other species, depending on the patrilineal or matrilineal subgenome, respectively, of the ancestral duplication of Cyprininae (4Rc). Chromosome coding corresponds to that of the genome project (Supplementary Table S2), and the (+) or (−) symbols represent the sense or antisense orientation of the chromosome, respectively, in the projects analyzed. Genes are represented with pentagons whose sharp vertex indicates the transcription sense. Same colors indicate orthologous genes. Black pentagons represent unknown genes or genes without orthologous in any chromosome analyzed. Mutations are inferred comparing the orthologous chromosomes: pseudogene (Ψ), deletion (#), tandem duplication (green dashed boxes), and inversion (red dashed boxes). Gene abbreviations are indicated in the Supplementary Table S3. Gene names in bold blue indicate diagnostic markers of patrilineal chromosomes.
|

